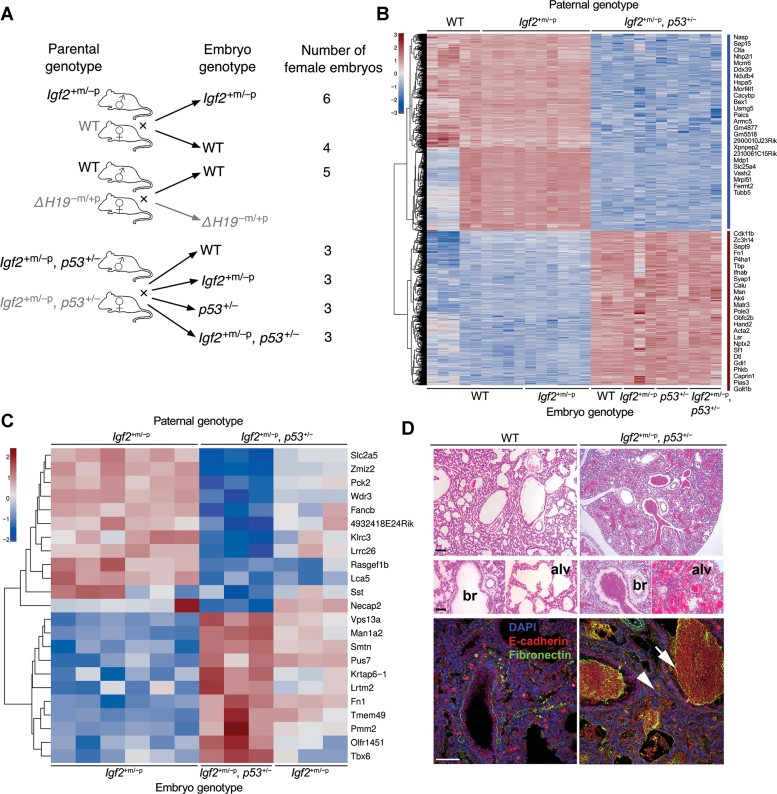
Schema of embryonic RNA expression analysis. Three parental genotype pairs were crossed to yield a number of female embryos of specific genotype as illustrated.
Comparison of gene expression in E9.5 female embryos derived from Igf2+m/−p or WT fathers (WT and Igf2+m/−p) with the expression in embryos from Igf2+m/−p, p53+/− fathers (WT, Igf2+m/−p, p53+/− and Igf2+m/−p, p53+/−). 1461 genes were significantly different in expression by SAM analysis and varied more than twofold, with most differentially expressed genes displayed in the heatmap.
Comparison of gene expression in Igf2+m/−p female embryos derived from WT fathers with those Igf2+m/−p and Igf2+m/−p, p53+/−embryos from Igf2+m/−p, p53+/− fathers. ‘Rescue’ signature of 23 genes shown in the heatmap.
H & E-stained lungs from E19.5 Igf2+m/−p female embryos (low power above, bar 1 mm, high power 100 µm) were filled with blood in both bronchioles (br) alveolar air-spaces (alv) when compared to WT littermates. Confocal images showed there were no gross differences in lung structure labelled with E-cadherin, but fibronectin was predominantly present in blood within the airspaces (white arrow), and subjectively increased in small airway and alveolar regions (white arrow head). Bar = 100 µm.