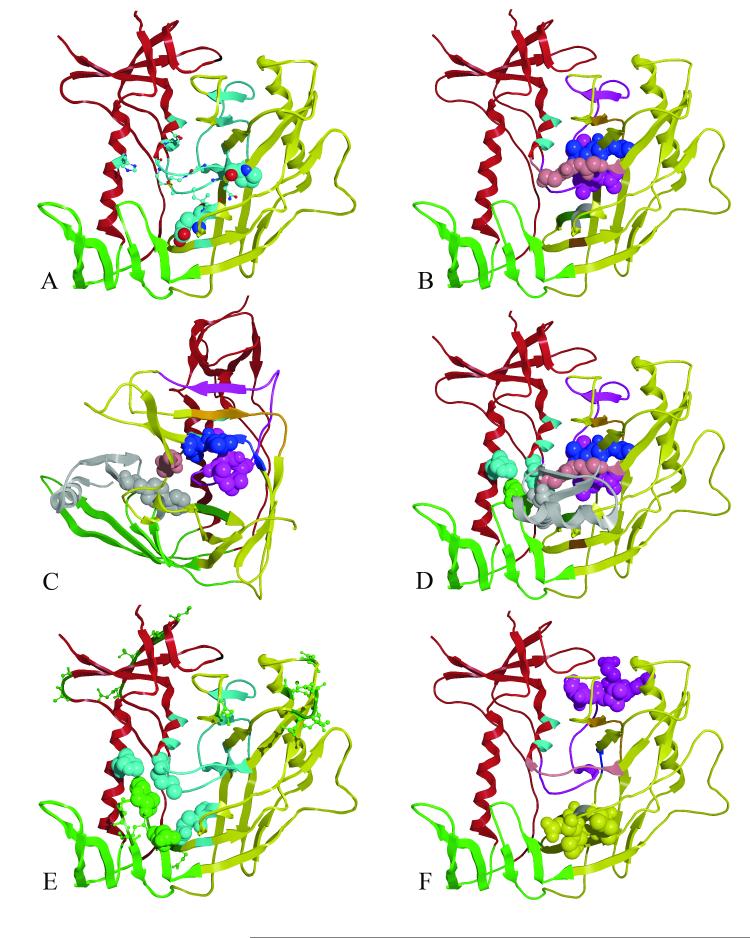
Figure 4. GNM slow mode minima and maxima, mapped to the gp120 core structure.
(A)The core gp120 inner (red), outer (yellow) and bridging sheets (green) domains are depicted as a ribbon diagram for GPO3 (pdb code, 2I5Y). Residues that have GNM minima are colored light blue. Residues with GNM minima which are conserved in all HIV gp120s are shown in spacing filling model (E370 and P470) and residues conserved in human HIV gp120s are shown as ball-and-stick (Q258, I371, G472, G473, M475, D477). ( B) Residues 252-275(purple) span the inner and outer domain and form the largest continuous stretch of GNM slow mode minima with the tripeptide L259-L260-L261 shown as space filling model. Adjacent GNM slow mode minima, L452, L453, L454: G471, G472, G473: I285, V286, Q287, L288, N289, E280, S291, S37 F376, N377 and P370, I371 are colored blue, pink, orange, dark green and gray respectively. (C) As in B but rotated 90 degrees with surface structural elements removed to depict proximity of the core GNM slow mode folding nucleus proximal to the 2I5Y mini-protein, biphenyl (gray) bound in the Phe 43 cavity. (D) GNM slow mode minima in the inner domain H105 (cyan) and M475 (cyan) are shown straddling W427 (green space filling model). (E) GNM slow mode minima and maxima are show in cyan and green ball and stick, respectively. Residues W427 and N425 (space filling green) are conserved residues locating on the bridging sheet. These GNM maxima likely for the recognition features that for ligand binding and interact with GNM minima (residues H105, M475, and E370, light blue space filling) which provide binding site stability. F) Residues E268, E269, I272, R273 and S274 (purple space filling model) have correlated GNM fluctuations with residues D368, P369, V372, T373 and S375 (yellow space filling model) that are more that 10.0 Å apart. (Rendered with MOE70)