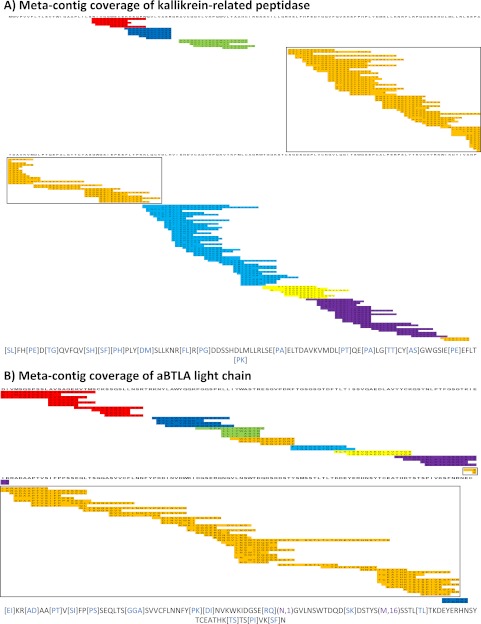
Fig. 4.
Mapped Meta-contigs. Meta-contig PRM spectra were aligned to reference proteins to evaluate de novo sequencing coverage. Every colored row corresponds to a contig PRM spectrum as separately mapped to the target protein sequence (information not used by Meta-SPS). Every set of overlapping contigs of the same color corresponds to a meta-contig; sets of contigs of the same color with no overlap indicate separate meta-contigs. Below each coverage map is the longest meta-contig sequence of the boxed meta-contig for the corresponding protein. Purple gaps correspond to mapped sequence calls with PTMs verified by MS-GFDB; blue gaps correspond to mapped gaps that span 2 or more residues in the reference. Remaining un-colored residues represent sequence calls that map to reference amino acid masses. A, Meta-contig coverage of kallikrein-related peptidase from the 6-prot sample is displayed here; 8 meta-contigs covered 78% of the 261 AA protein with the longest sequence spanning 94 AA. B, Meta-contig coverage of the aBTLA light chain is displayed here; 9 meta-contigs covered 87% of the 219 AA protein with the longest sequence spanning 107 AA.