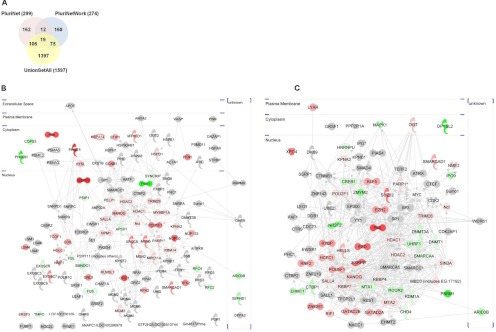
Fig. 3.
Intersecting proteomics studies with existing pluripotency networks. A, Overall overlap of all identified proteins in union set of proteins identified from gel-free and gel-based proteomics studies (“union set all” - 2 peptides; Scaffold probability ≥ 99%) with PluriNet and PluriNetWork. B, The intersection between proteomics data and PluriNet (analyzed using Ingenuity Pathways Analysis) - 125 proteins. C, The intersection between proteomics data and PluriNetwork - 94 proteins. Each node in (B) and (C) represents a protein identified in mESC or mEpiSC samples (red shaded nodes indicate mESC>mEpiSC protein expression with p < 0.1 (Student's t test), green shaded indicate mEpiSC>mESC protein expression with p < 0.1 (Student's t test) and gray shaded nodes indicate identified protein in either mESC or mEpiSC but without statistically significant quantitative difference.