Abstract
Post-translational modification of proteins by lysine acetylation plays important regulatory roles in living cells. The budding yeast Saccharomyces cerevisiae is a widely used unicellular eukaryotic model organism in biomedical research. S. cerevisiae contains several evolutionary conserved lysine acetyltransferases and deacetylases. However, only a few dozen acetylation sites in S. cerevisiae are known, presenting a major obstacle for further understanding the regulatory roles of acetylation in this organism. Here we use high resolution mass spectrometry to identify about 4000 lysine acetylation sites in S. cerevisiae. Acetylated proteins are implicated in the regulation of diverse cytoplasmic and nuclear processes including chromatin organization, mitochondrial metabolism, and protein synthesis. Bioinformatic analysis of yeast acetylation sites shows that acetylated lysines are significantly more conserved compared with nonacetylated lysines. A large fraction of the conserved acetylation sites are present on proteins involved in cellular metabolism, protein synthesis, and protein folding. Furthermore, quantification of the Rpd3-regulated acetylation sites identified several previously known, as well as new putative substrates of this deacetylase. Rpd3 deficiency increased acetylation of the SAGA (Spt-Ada-Gcn5-Acetyltransferase) complex subunit Sgf73 on K33. This acetylation site is located within a critical regulatory domain in Sgf73 that interacts with Ubp8 and is involved in the activation of the Ubp8-containing histone H2B deubiquitylase complex. Our data provides the first global survey of acetylation in budding yeast, and suggests a wide-ranging regulatory scope of this modification. The provided dataset may serve as an important resource for the functional analysis of lysine acetylation in eukaryotes.
Lysine acetylation is a dynamic and reversible post-translational modification. Acetylation of lysines on their ε-amino group is catalyzed by lysine acetyltransferases (KATs1, also known as histone acetyltrasferases (HATs)), and reversed by lysine deacetylases (KDACs, also known as histone deacetylases (HDACs)) (1). The enzymatic machinery involved in lysine acetylation is evolutionary conserved in all forms of life (2–4). The role of acetylation has been extensively studied in the regulation of gene expression via modification of histones (5). Acetylation also plays important roles in controlling cellular metabolism (6–10), protein folding (11), and sister chromatid cohesion (12). Furthermore, acetylation has been implicated in regulating the beneficial effects of calorie restriction (13), a low nutrient diet without starvation, and aging. Based on these findings, it is proposed that the functional roles of acetylation in these processes are evolutionary conserved from yeast to mammals.
Advancements in mass spectrometry (MS)-based proteomics have greatly facilitated identification of thousands of post-translational modification (PTM) sites in eukaryotic cells (14–18). Proteome-wide mapping of PTM sites can provide important leads for analyzing the functional relevance of individual sites and a systems-wide view of the regulatory scope of post-translational modifications. Also, large-scale PTM datasets are an important resource for the in silico analysis of PTMs, which can broaden the understanding of modification site properties and their evolutionary trajectories.
The budding yeast Saccharomyces cerevisiae is a commonly used unicellular eukaryotic model organism. Yeast has been used in many pioneering “-omics” studies, including sequencing of the first eukaryotic genome (19), systems-wide genetic interactions analysis (20, 21), MS-based comprehensive mapping of a eukaryotic proteome (22), and proteome-wide analysis of protein-protein interactions (23, 24). In addition, S. cerevisiae has been extensively used to study the molecular mechanisms of acetylation. Many lysine acetyltransferases and deacetylases were discovered in this organism (2, 25), and their orthologs were subsequently identified in higher eukaryotes. Furthermore, the functional roles of many well-studied acetylation sites on histones are conserved from yeast to mammals. Recent data from human and Drosophila cells show that acetylation is present on many highly conserved metabolic enzymes (26–28). However, only a few dozen yeast acetylation sites are annotated in the Uniprot database. Given the presence of a well-conserved and elaborate acetylation machinery in yeast, we reasoned that many more acetylation sites exist in this organism that remained to be identified.
Here we used high resolution mass spectrometry-based proteomics to investigate the scope of acetylation in S. cerevisiae. We identified about 4000 unique acetylation sites in this important model organism. Bioinformatic analysis of yeast acetylation sites and comparison with previously identified human and Drosophila acetylation sites indicates that many acetylation sites are evolutionary conserved. Furthermore, quantitative analysis of the Rpd3-regulated acetylation sites identified several nuclear proteins that showed increased acetylation in rpd3 knockout cells. Our results provide a systems-wide view of acetylation in budding yeast, and a rich dataset for functional analysis of acetylation sites in this organism.
MATERIALS AND METHODS
Yeast Cultures and Preparation of Protein Lysates
S. cerevisiae cells from a lysine auxotroph strain (BY4742) were grown in a synthetic complete medium containing “light” lysine (l-lysine 12C6,14N2, or Lysine0) or “heavy” lysine (l-lysine 13C6,15N2, or Lysine8) for more than 10 generations to an absorbance (measured at 600 nm (OD600)) value of ∼0.7. For quantification of the Rpd3-regulated acetylation sites, we used a lysine auxotroph rpd3 deletion strain (rpd3Δ) obtained from the MATα yeast knockout collection (29). The wild-type control strain (BY4742) was grown in media supplemented with “light” lysine whereas rpd3Δ cells were cultured in “heavy” lysine containing media, to an OD600 of ∼0.5. Cells were harvested by centrifugation at 3000 × g for 5 min, washed once in water, and resuspended in lysis buffer (150 mm KAc, 2 mm MgAc, 20 mm Hepes, pH 7.4) supplemented with Protease Inhibitor Mixture (Roche). The cell-suspension was instantly frozen in liquid nitrogen and cells were cryo-grinded using a MM400 ball mill (Retsch) for 2 × 3 min at 25 Hz. Frozen powdered lysates were transferred to 50 ml tubes, thawed, and Triton-X was added to a final concentration of 1%, followed by incubation at 4 °C for 30 min. Extracts were centrifuged twice at 3500 × g for 10 min to remove cellular debris. Protein concentrations were measured using BCA protein assay kit (Thermo Scientific) and lysates were stored at −80 °C until further use.
MS Sample Preparation
Cell lysates were thawed and 20 mg of proteins from “light” and “heavy” labeled samples were mixed. Proteins were precipitated with 4 volumes of ice-cold acetone. After incubation at −20 °C for more than 1 h, the precipitate was pelleted at 3000 × g for 10 min and the liquid phase was discarded. The protein pellet was dissolved in 8 m urea solution (6 m urea, 2 m thiourea, 10 mm HEPES pH 8.0) and the protein concentration was measured using Bradford assay (BioRad, Hercules, CA). Protein samples were treated with 1 mm DTT for 45 min and subsequently alkylated with 5.5 mm iodoacetamide for 45 min in the dark. Proteins were digested with endoproteinase Lys-C (1 μg Lys-C per 100 μg protein) for 12–16 h at 25 °C. Samples were subsequently diluted with H2O to reduce the urea concentration to 2 m and the pH was adjusted to 8.0 with 1 m ammonium bicarbonate solution. Trypsin endoproteinase was added (1 μg Trypsin per 100 μg protein) and the samples were further digested for 12–16 h at 25 °C. Trypsin activity was stopped by addition of trifluoroacetic acid (TFA) to a final concentration of 1% and the peptide solution was incubated at 4 °C for 1–2 h. The solution was clarified by centrifugation at 3000 × g and peptides were purified using a C18 Sep-Pak cartridges (Waters, Milford, MA). Peptides were eluted from the C18 cartridges with 5 × 2 ml of 40% acetonitrile, 0.1% TFA in H2O. To remove acetonitrile, the sample was freeze-dried. Peptides were redissolved in immune affinity purification buffer (50 mm 3-(N-morpholino)propanesulfonic acid pH 7.2, 10 mm Na3PO4, 50 mm NaCl) and the peptide solution was clarified by centrifugation at 13000 × g for 5 min. Fifty microliters of agarose conjugated anti-acetyllysine antibodies (ImmuneChem Pharmaceuticals Inc., Burnaby, Canada, catalog number ICP0388) were added to the peptide mix (∼20 mg) and incubated on a rotating wheel at 4 °C for 12–16 h. The antibody batch numbers 022808 and 091211 were used for obtaining data set 1 (supplemental Table S1), and dataset 2 (supplemental Table S2), respectively. The immunoprecipitates were washed 3 times with ∼20 volumes ice-cold immunoaffinity purification (IAP) buffer and subsequently three times with ∼20 volumes ice-cold H2O. Peptides were eluted with 0.15% TFA in water. The eluted peptides were separated into 12 fractions by isoelectric focusing as described (30, 31), or fractioned into four fractions by micro-column-based strong cation exchange fractionation (32). The fractionated peptides were purified on StageTips packed with C18 reversed-phase material (32).
MS Analysis
Peptide fractions from immunoprecipitated samples were analyzed on a hybrid linear ion-trap Orbitrap (LTQ-Orbitrap Velos, Thermo Scientific) (33) or quadrupole Orbitrap (Q-Exactive, Thermo Scientific) mass spectrometer (34, 35) equipped with a nanoflow HPLC system (Thermo Scientific) as described. Peptide samples were loaded onto C18 reversed phase columns (15 cm length, 75 μm inner diameter) and peptides were eluted with a linear gradient (3–4 h) from 8 to 40% acetonitrile containing 0.5% acetic acid. The mass spectrometers were operated in data dependent mode, automatically switching between MS and MS2 acquisition. Survey full scan MS spectra (m/z 300–1700) were acquired in the Orbitrap. The 10 most intense ions were sequentially isolated and fragmented by higher-energy C-trap dissociation (HCD) (36). An ion selection threshold of 5000 counts was used. Peptides with unassigned charge states, as well as with charge states less than +2 were excluded from fragmentation. Fragment spectra were recorded in the Orbitrap mass analyzer. A lock mass ion from ambient air (m/z 445.120025) was used for internal calibration of measurements in the Orbitrap on LTQ-Orbitrap Velos mass spectrometers (37).
MS-data Processing
Raw MS data files obtained from the LTQ Orbitrap Velos or Q-Exactive were processed using MaxQuant (development version 1.2.7.1, http://www.maxquant.org/) (38, 39). Peptide ion masses and fragment spectra were searched against the Saccharomyces Genome Database (SGD) (40) release 63 containing 6717 putative protein sequences (http://downloads.yeastgenome.org/). Peptide sequences were searched using trypsin specificity and allowing a maximum of two missed cleavages. Carbamidomethylation of cystein was searched as fixed modification and methionine oxidation, N-terminal acetylation, and lysine acetylation were added as variable modifications. MaxQuant presearch determined spectra that result from heavy stable isotope labeling by amino acids in cell culture (SILAC) labeled peptides and searched these with the additional fixed modification of Lys8, whereas spectra with a SILAC state not determined in presearch were searched with Lys8 as additional variable modification. Lysine acetylation sites were required to be located internally within modified peptide sequences. Database searching was performed with 6 ppm mass tolerance for precursor ions and 20 ppm for fragment ions. The false discovery rate (FDR) was estimated using a target-decoy approach (41) allowing a maximum of 1% false identifications from a reversed sequence database. Localization probabilities were calculated by MaxQuant as previously described (42). MS2 spectra associated with acetylated peptides reported here can be found in supplemental Fig. S3 and S4.
Bioinformatic Analysis
Statistical analysis was performed using the R software environment. Analysis of the amino acid sequences surrounding acetylation sites was performed using iceLogo (43). Evolutionary conservation analysis was performed using orthology assignments and multi-sequence alignments from the evolutionary genealogy of genes: nonsupervised orthology groups (eggNOG) database version 2.0 (44). The eggNOG database contains extended versions of the manually curated eurkaryotic orthology groups (KOGs) and adds additional eukaryotic nonsupervised orthology groups (euNOGs), thereby providing a broad coverage of protein sequences over different species. First, all modified yeast peptide sequences were mapped to eggNOG protein sequences and orthology groups. Only peptides matching to sequences in a single orthology group were considered for further analysis to avoid over-counting of ambiguous peptide matches. Lysine conservation was determined for each species and at each alignment position that corresponds to a yeast modification site separately. A lysine residue was considered to be conserved if at least one sequence in the multi-sequence alignment contained a lysine residue at the aligned position. A dataset containing all lysine residues of all proteins identified in this study served as control. Mean conservation of modified residues and control residues were plotted separately for each species. p values were separately calculated for each species using Fisher's exact test.
Gene Ontology (GO) annotation analysis was performed using the GO Slim Mapper from the SGD. p values were calculated separately for each GO slim term using Fisher's exact test. A p value < 0.01 (adjusted for multiple comparisons) was considered statistically significant. Alternatively, gene ontology biological process term enrichment analysis was performed using the DAVID bioinformatics resource (45).
Functional interaction network analysis was performed using interaction data from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (46). Only high confidence interactions (score > 0.7) are represented in the networks. Interactions derived from text-mining were excluded. Cytoscape (47) version 2.8 was used for visualization of protein interaction networks.
Computational Modeling and Simulations
The initial model of Ubp8-Sgf11-Sgf73-Sus1 SAGA DUB module complex was prepared from the available crystal structure (PDB code 3M99) (48). The missing regions/loops in the crystal structure were modeled using modeler (49) via the MODWEB (http://salilab.org/modweb) and MODLOOP (http://salilab.org/modloop) servers. For acetylated K33 (Ac-K33) of the Sgf73 domain, the lysine residue was modified using Sirius visualization system. The protein complex was then solvated in simple point charge (SPC) water molecules in a dodecahedron box, with the box edges ∼1.0 nm from any atom of the protein. Appropriate numbers of sodium ions were added to maintain system neutrality. The system was energy minimized using steepest descent and after an equilibration of 200 ps, the system was subjected to 5 ns of molecular dynamics (MD) simulation. All MD simulations were carried out with the GROMACS 4.0.7 simulation software package (50) and the GROMOS96 43a2 force field (51). The simulations were performed under a constant temperature of 300 K using a coupling constant of 0.1 ps and constant pressure of 1 atm with a coupling constant of 1.0 ps. The V-rescale algorithm (52) was used for temperature and pressure was maintained using Parrinello-Rahman method (53). Van der Waals interactions used a simple cut off at 1.2 nm and long range electrostatics were handled using the Fast Particle-Mesh Ewald method (PME) (54) with a cutoff of 1.2 nm. All bonds were constrained using the LINCS algorithm (55). The water molecules were constrained using SETTLE algorithm (56). The time step used in the simulations was 2 fs.
RESULTS
Proteome-wide Mapping of Lysine Acetylation Sites in S. cerevisiae
To map lysine acetylation sites in budding yeast we used proteins prepared from exponentially growing (OD600 0.7) S. cerevisiae cells. Proteins were digested into peptides using trypsin, and acetylated peptides were immuno-enriched using anti-acetyllysine antibodies as described previously (26, 57) (Fig. 1A). To identify low abundant modification sites, the sample complexity was further reduced by fractionating peptides using isoelectric focusing (30, 31). Peptides were analyzed with liquid chromatography coupled to a high resolution Orbitrap mass spectrometer (LTQ Orbitrap Velos or Q Exactive). Peptides were fragmented using HCD (36) and intact peptide ions as well as their fragment ion masses were measured in the Orbitrap mass analyzer. Raw data were processed using MaxQuant (38), and mass spectra were searched using the Andromeda search engine (58).
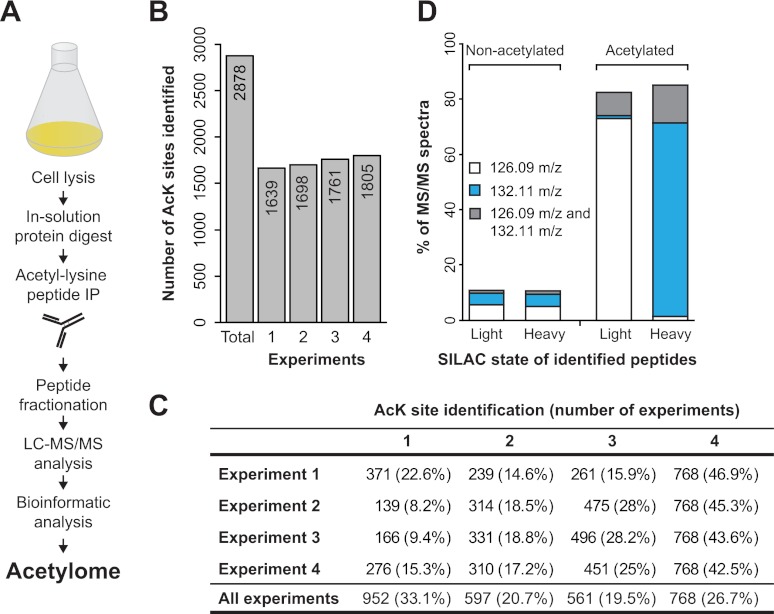
Fig. 1.
Proteome-wide identification of lysine acetylation sites in S. cerevisiae. A, Schematic representation of the workflow used for the identification of yeast acetylation sites. Yeast cells were harvested and mechanically lysed, and proteins were digested into peptides and acetylated peptides were enriched using anti-acetyllysine antibodies. The enriched peptides were further fractionated by isoelectric focusing and analyzed by mass spectrometry. B, The graph shows the number of acetylation sites identified in each of four experiments, as well as total number of acetylation sites identified in all four experiments. The number of identified acetylation sites is indicated within each bar. C, The table shows the number of acetylation sites identified in each experiment and the percent of acetylation sites identified in one or more experiments. For example, for any of four experiments 77–92% of sites were identified in more than one experiment. D, The presence of the acetyllysine diagnostic ion in MS fragment scans of acetylated peptides and nonacetylated peptides. The mass of the diagnostic ion (126.09 m/z or 132.11 m/z) is closely associated with the SILAC state of the lysine from corresponding fragment scans.
To obtain an in-depth view of the lysine acetylation in yeast we performed four biological replicate experiments. Forty-eight samples (12 fractions from each experiment) were analyzed by liquid chromatography coupled to electrospray ionization mass spectrometry (LC-MS/MS). Data from all four experiments were processed collectively allowing a FDR of 1% at both peptide and site level. Assuming that trypsin fails to cleave at the C terminus of acetylated lysines, we required that all acetylated peptides should contain at least one lysine that is internal to the peptide sequence. After applying these parameters, we identified 2,878 acetylation sites (supplemental Table S1) that matched to 1,059 unique yeast open reading frames (ORFs) from the Saccharomyces Genome Database (SGD) (40). Acetylation of four randomly selected proteins was verified by anti-acetyllysine immunoblotting (supplemental Fig. S1A). To assess the reproducibility of our experimental approach, we calculated the overlap of identified acetylation sites between the four experiments. A majority (∼67%) of the reported acetylation sites were identified in two or more replicate experiments and about 77–92% of the acetylation sites identified in any one experiment were found in at least one other experiment (Figs. 1B and 1C).
Evaluation of the Acetyllysine Diagnostic Ion in the Identification of Acetylation Sites
It has been reported that fragmentation of acetyl-lysine containing peptides can yield a highly specific “diagnostic” ion at m/z 126 (precise mass 126.0913 Da) (59), which can provide additional confidence for the presence of acetylated lysine within a peptide. This marker ion is derived from an acetyl-lysine immonium ion (Im) by loss of ammonia, here referred to as Im(acK). The presence of this diagnostic ion was previously assessed using synthetic peptides or in vitro chemically acetylated peptides (59, 60). However, its utility has not been tested in a large-scale in vivo acetylation analysis. To date, the majority of MS-based large-scale acetylation analyses have been performed using the collision induced dissociation (CID) fragmentation method where fragment ions were detected in a linear ion trap. However, the ion trap mass analyzers suffer from the so called “one-third cut off rule” where peptide fragment ion masses lower than one-third of their intact peptide ion mass cannot be observed in the MS/MS spectra. The Im(acK) ion is present in the low mass region of MS/MS spectra and is frequently not recorded in CID spectra. Thus, a significant advantage of HCD fragmentation is that it enables measurement of peptide fragment ions in the low mass region with high precision. To test the specificity of the diagnostic Im(acK) ion, we scanned the MS/MS spectra of all peptides sequenced with the HCD method. We performed all our experiments using SILAC by growing yeast cells in media containing lysine0 or lysine8 (61). In a SILAC experiment, the mass of the Im(acK) ion depends on the SILAC state of the precursor peptide (for lysine0 the diagnostic ion mass is: 126.0913 Da; for lysine8 the diagnostic ion mass is: 132.1050 Da). With this experimental setup we were able to investigate the diagnostic relevance of this ion for acetylation site identifications. In our dataset we find that a large fraction (>80%) of the fragment spectra of acetylated peptides contains the Im(acK) ion (Fig. 1D). As expected, we find that the m/z of Im(acK) ion is tightly associated with the SILAC state of the sequenced peptides—the light ion (126.0913 Da) is found in fragment scans of light SILAC peptides, whereas the heavy ion (132.1050 Da) is found in fragment scans of heavy SILAC peptides. Although the presence of the Im(acK) ion appears to correlate well with the acetylated peptides in our data, we note that the ion is also present in a small fraction of MS/MS scans assigned to unmodified peptides. In these spectra associated with nonacetylated peptides the mass of the diagnostic ion did not correlate with the SILAC state of the identified peptide, indicating that the Im(acK) ion could be derived from co-eluting peptides. To confirm the specificity of the Im(acK) ion, we performed an acetylation analysis from non-SILAC samples. In these data we only find the Im(acK) ion with a mass of 126.0913 Da in the spectra of acetylated peptides, and no MS/MS spectra containing a mass of 132.1050 Da (data not shown). These results confirm that the Im(acK) diagnostic ion is highly specific for acetylation. However, it also indicates that the diagnostic value of the Im(acK) ion for individual site identifications in large-scale acetylation studies is somewhat limited. In retrospect, this is not surprising because the selection window for precursor ions is several mass units wide and often admits a fraction of co-eluting peptides (30, 31).
Distribution of Acetylation Sites in the S. cerevisiae Proteome
To assess the distribution of acetylation sites in the yeast proteome, we calculated the number of identified modification sites per protein. Although about 50% of proteins contained only one acetylation site, many other proteins in our dataset were acetylated on multiple lysines (Fig. 2A). Among the highly acetylated proteins are many mitochondrial enzymes that are involved in diverse metabolic pathways (supplemental Table S1). Identification of PTMs on low abundant proteins can be challenging in MS-based studies because of a limited dynamic range of the instruments. We used the known abundance distribution of yeast proteins that was estimated by measuring copy numbers of green fluorescent protein (GFP) -tagged yeast ORFs (62) to compare the abundance of acetylated proteins identified in our study with the abundance of all yeast proteins (Fig. 2B). The results obtained from this analysis indicate a bias for detecting acetylation sites on more abundant proteins, and suggest that our dataset is unlikely to be comprehensive. Regardless, the number of acetylation sites identified in yeast is comparable to the number of sites identified in human and Drosophila, suggesting that acetylation is as widespread in yeast as in metazoans.
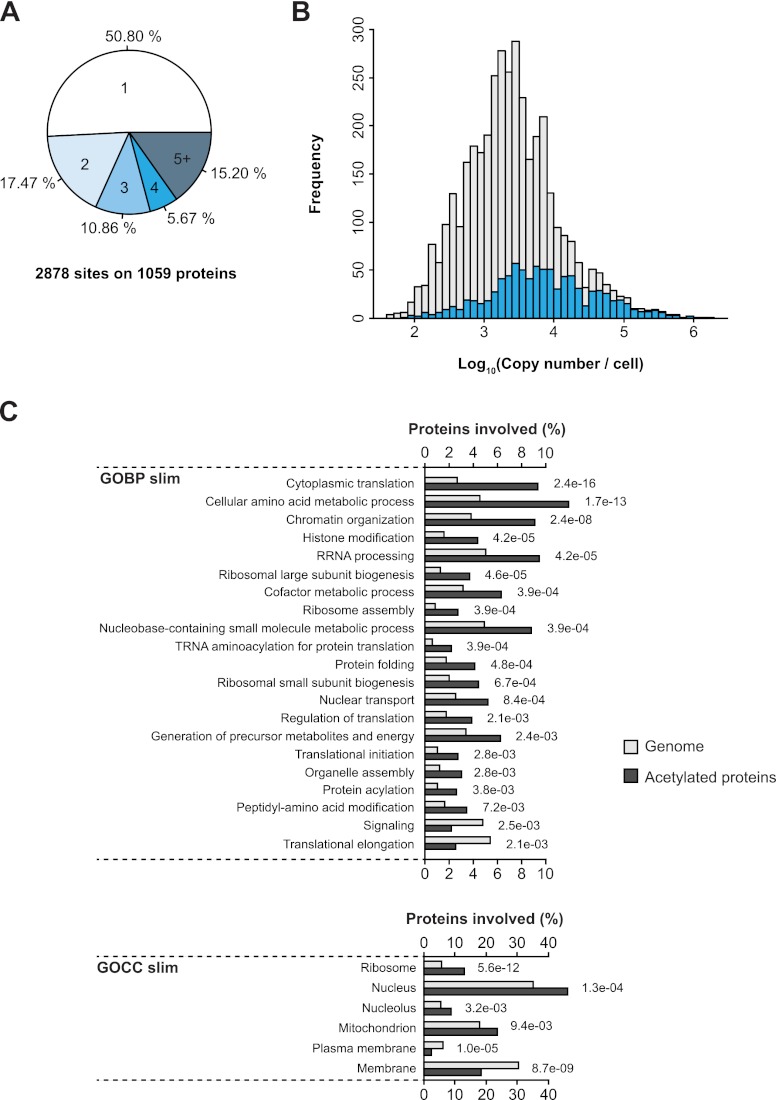
Fig. 2.
Acetylation targets proteins involved in diverse biological processes. A, The pie chart illustrates the number of lysine acetylation sites identified per protein. B, The abundance distribution of acetylated proteins. The abundance of acetylated proteins was compared with the abundance of all cellular proteins. C, Comparative GO slim biological process and cellular compartment term enrichment analysis of acetylated proteins and all yeast proteins.
Cellular Localization and Functional Annotations of Acetylated Proteins
Gene Ontology (GO) enrichment analysis was performed on acetylated proteins to examine the preferential subcellular localization of acetylated proteins and to reveal the biological processes they are involved in. Acetylated proteins were more frequently present in the cytoplasm, nucleus, and mitochondria whereas they were underrepresented in the membrane compartments (Fig. 2C). Several GOBP (GO biological process) terms were significantly overrepresented in our data, including enzymes involved in amino acid metabolism, histone modification, ribosome assembly, and terms related to translational control. The role of acetylation in histone modification and its effects on control of gene expression has been extensively studied over the last decades. We find that acetylation occurs on many proteins involved in diverse nuclear processes such as RNA processing, DNA repair, and nuclear transport suggesting a broad regulatory scope of this modification in the nucleus. Interestingly, we find that a sizable portion of acetylation sites is located on cytoplasmic proteins, including proteins involved in protein synthesis and protein folding. Many of these proteins were previously found to be acetylated in human and Drosophila cells (26, 28), implying that acetylation may play an evolutionary conserved role in regulating protein synthesis.
Acetylation of Metabolic Enzymes
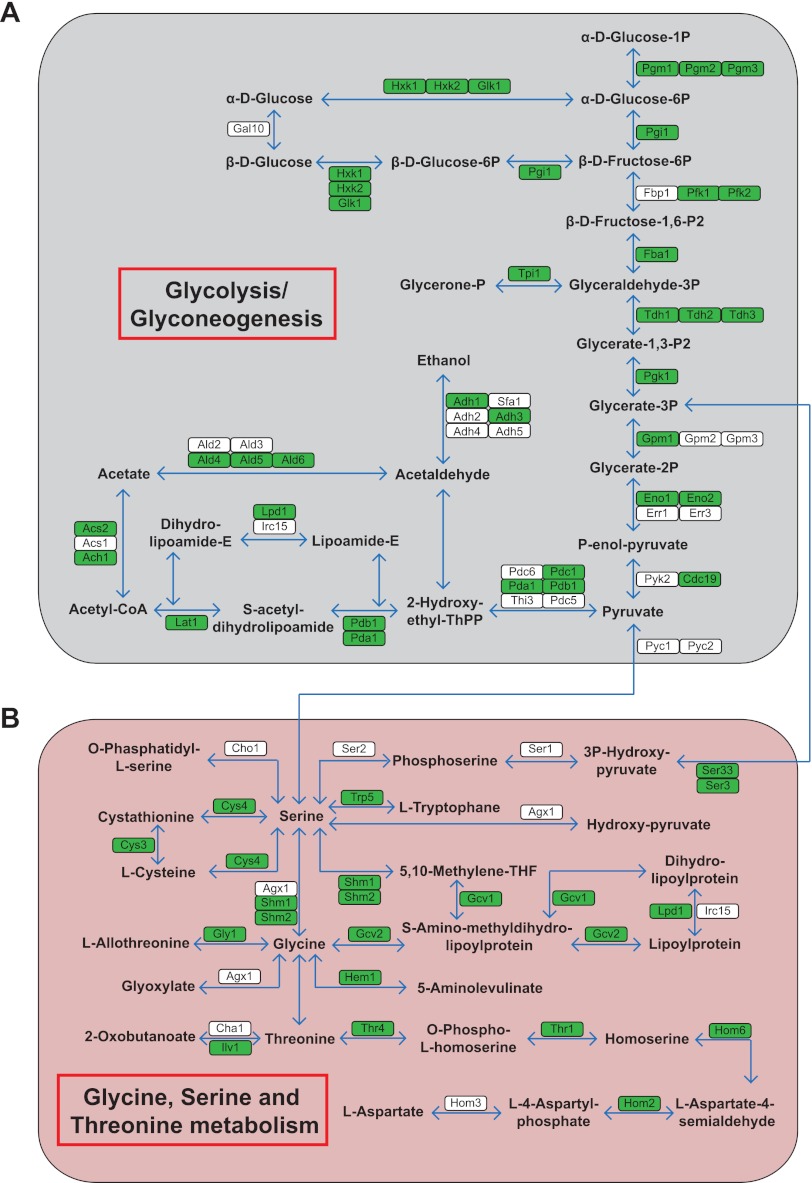
The enzymes involved in central metabolic pathways are highly conserved, and many acetylation sites have been identified in human (26, 27), Drosophila (28), and bacterial metabolic enzymes (63–65). We investigated the acetylation of metabolic enzymes in yeast by mapping acetylated proteins to KEGG metabolic pathways (66). A large proportion of metabolic enzymes that are involved in glycolysis/gluconeogenesis and amino acid metabolisms, such as the glycine/serine/threonine metabolism pathway are acetylated (Fig. 3). Several enzymes involved in these pathways were previously reported as acetylated in various organisms (26, 28, 63, 65). The fact that nearly all enzymes in major metabolic pathways are subjected to acetylation implies that this modification may be intricately linked to cellular metabolism at multiple levels. Hence, our results support the notion that acetylation may have an anciently conserved role in controlling cellular metabolism (7). However, given that many of these proteins are acetylated on multiple lysines, deciphering their functional roles may be a challenging task.
Fig. 3.
Acetylation of metabolic enzymes in yeast. Schematic representation of the enzymes involved in Glycolysis/Glyconeogenesis (A) and glycine, serine, and threonine metabolism (B). A majority of the enzymes involved in these biological processes were identified to be acetylated (color-coded in green).
Evolutionary Conservation of Acetylated Lysines
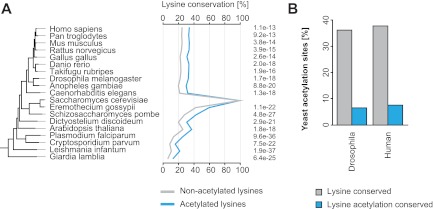
Several previously known acetylation sites in yeast proteins such as histones (67, 68), SAS2 (69), and SMC3 (12, 70, 71), are conserved in diverse eukaryotic organisms. To evaluate the degree of conservation of acetylated lysines identified in our screen, we performed a bioinformatic analysis using the evolutionary genealogy of genes: eggNOG database (44). Orthologs of acetylated yeast proteins were obtained for diverse prokaryotic and eukaryotic organisms, and the conservation of acetylated lysines was determined by aligning protein sequences of yeast with orthologs from the other indicated species. These analyses show that in comparison to nonacetylated lysines, acetylated lysines in yeast are significantly more conserved (Fig. 4A). Furthermore, we used previously reported human (26) and Drosophila acetylation sites (28) to analyze the extent of site-specific conservation of acetylation. About one-third of acetylated lysines were found to be conserved between these species, and for a fraction of this acetylation was detected in Drosophila and human cells (Fig. 4B).
Fig. 4.
Acetylation sites show a high degree of evolutionary conservation. A, Lysine conservation of yeast acetylated lysines in other species. Acetylated lysines show a significantly higher conservation than nonacetylated lysines. B, Conservation of lysine and lysine acetylation from yeast to Drosophila and human.
Quantitative Analysis of the Rpd3-regulated Acetylation Sites
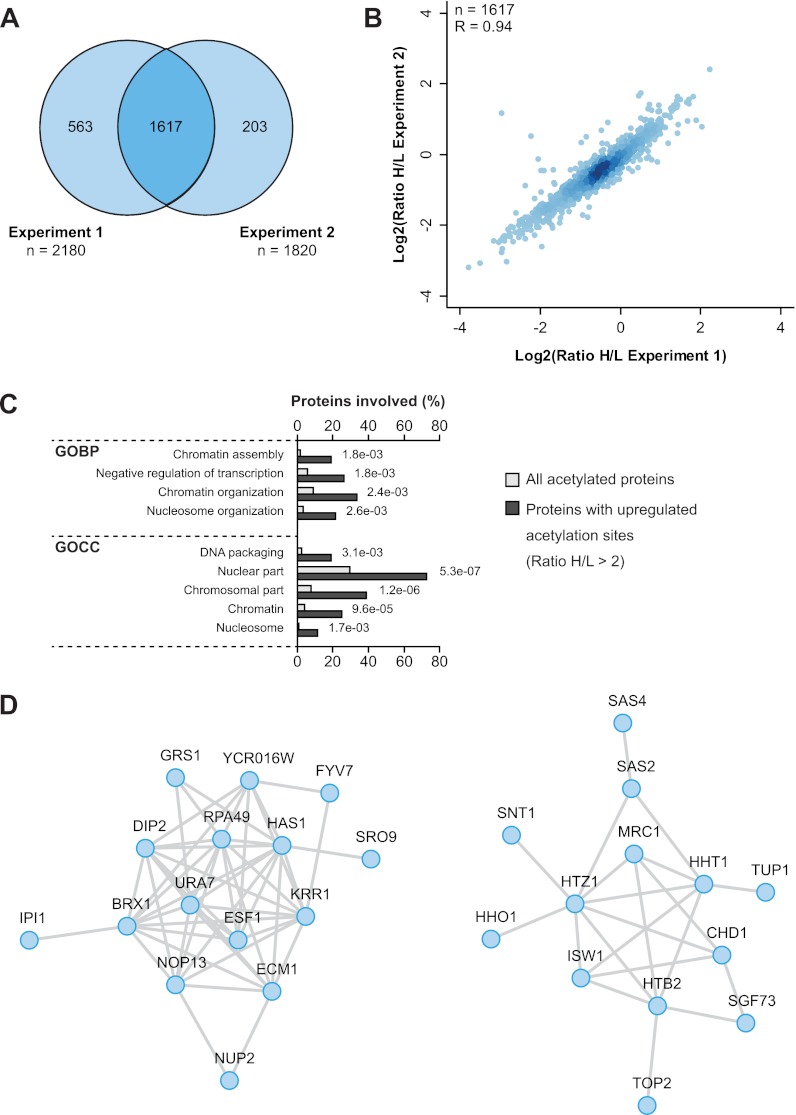
The enzymatic lysine deacetylation machinery is well-characterized in S. cerevisiae and consists of ten different KDACs. Among the class I KDACs Rpd3 is the most extensively studied deacetylase. Rpd3 is implicated in several biological processes, including regulation of histone acetylation (67, 72), transcriptional repression and chromatin silencing (73–75). To investigate the Rpd3-regulated acetylome, we applied a SILAC-based proteomics approach. Control wild-type (WT) cells were cultured in “light” SILAC media, and rpd3Δ cells were cultured in “heavy” SILAC media (supplemental Fig. S1B). Acetylated peptides were identified and quantified by mass spectrometry. In two independent biological replicates we identified over 2300 sites (Fig. 5A, supplemental Table S2). About two-thirds of the acetylation sites were quantified in both replicates with high experimental reproducibility (Fig. 5B).
Fig. 5.
Increased acetylation of nuclear proteins in rpd3Δ cells. A, The Venn diagram shows the overlap of acetylation sites quantified in two independent replicate experiments comparing acetylation in wild-type cells with rpd3 knockout cells. B, The scatter plot illustrates the experimental reproducibility of the two Rpd3 replicate experiments performed. Pearson's correlation coefficient is indicated. C, Gene Ontology enrichment analysis of proteins with increased acetylation (SILAC ratio >2) in rpd3Δ cells. GO terms related to nuclear processes showed significant enrichment. D, Functional interaction networks of proteins displaying increased acetylation in rpd3Δ cells.
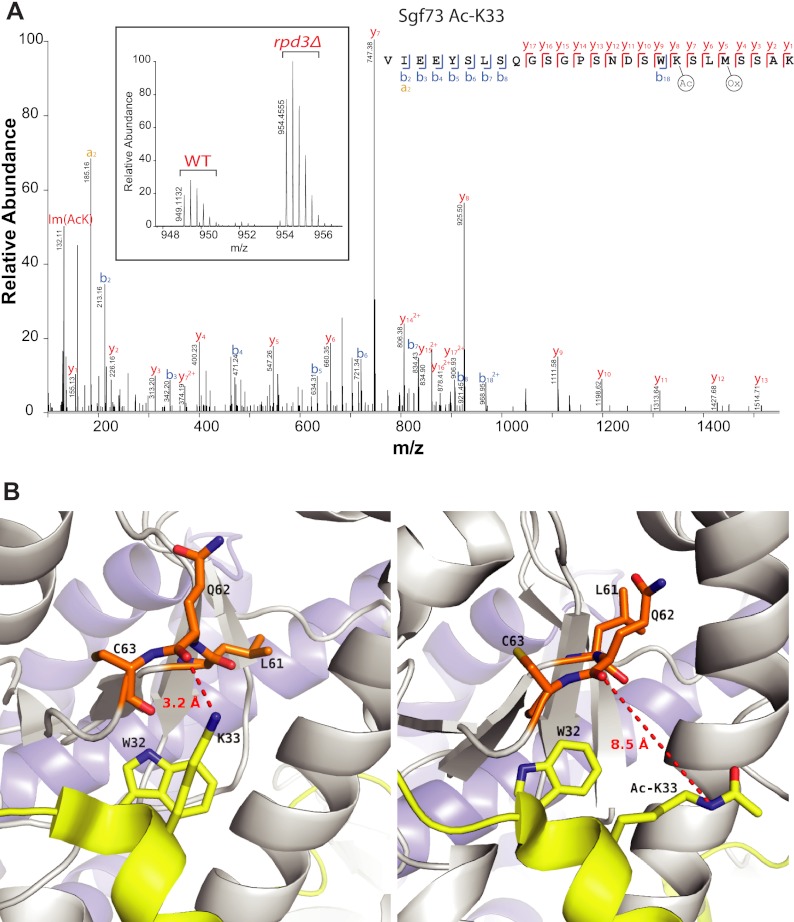
A Gene Ontology (GO) enrichment analysis of proteins with Rpd3-regulated acetylation sites showed enrichment of GO terms related to nuclear compartment and chromatin-associated processes (Fig. 5C). These data are in agreement with the known localization of Rpd3 to the nucleus and its previously reported function in regulating gene transcription. Among the Rpd3-regulated acetylation sites several are present on histones and subunits of histone acetyltransferases (supplemental Table S2). Many proteins that showed increased acetylation in rpd3Δ cells are functionally interconnected (Fig. 5D). Acetylation of the SAGA (Spt-Ada-Gcn5-A cetyltransferase) complex subunit Sgf73 is increased on lysine 33 (Fig. 6A). Interestingly, Lys33 of Sgf73 is located within the conserved WK motif which directly interacts with Ubp8 (48, 76). Ubp8 forms a deubiquitylase complex with the SAGA subunits that deubiquitylates histone H2B (77). The WK motif of Sgf73 forms direct hydrogen bonds with Ubp8 Q62 and mutations in the WK motif weakens the interaction between Sgf73 and Ubp8 (48). Computational modeling and simulation studies suggested that neutralization of the positive charge on K33 by acetylation places it further away from the side chain of Ubp8 Q62, which may weaken the interaction between Sgf73 and Ubp8 (Fig. 6B). This interaction is maintained throughout in the control simulation (data not shown).
Fig. 6.
The rpd3Δ cells show increased acetylation of Sgf73 on lysine 33. A, MS and MS/MS spectra of Sgf73 peptide containing acetylated lysine 33. The MS spectrum shows increased intensity of the peptide in rpd3Δ cells compared with wild-type control cells, and the fragment spectrum supports the peptide sequence identification and localization of the acetyl group on the indicated lysine. B, Lysine 33 of Sgf73 is located at the interaction interface of Sgf73 and Ubp8. The left panel shows cocrystal structure of Sgf73 and Ubp8 demonstrating a role of the K33 side chain in forming a hydrogen bond with the side chain of Q62 of Ubp8. The right panel shows a snapshot from the MD simulations, indicating that acetylation of Sgf73 within the WK motif at K33 neutralizes its positive charge, and moves it away from Upb8 Q62 side chain, with potential to weaken the interaction between Sgf73 and Ubp8.
Interestingly, a comparison of acetylation sites identified in our two datasets (dataset 1 from Fig. 1, supplemental Table S1; dataset 2 from Fig. 5, supplemental Table S2), showed that less than half of the acetylation were found in both experiments (supplemental Fig. S2A). Thus, combining the results from these two datasets resulted in the identification of about 4000 acetylation sites. To understand the reasons for the relatively low overlap between these two datasets, we compared the amino acid sequences flanking nonoverlapping sites from each dataset. These analyses revealed that nonoverlapping sites are biased for different sequences flanking the modified lysines (supplemental Fig. S2B). We used two different anti-acetyllysine antibody batches for these experiments, and these results may reflect batch-to-batch variability of these antibodies. This suggest that although antibody-based approaches are useful for identifying endogenous acetylation sites, caution should be taken in the interpretation of acetylation motifs as differences in antibody specificities may affect the outcome of such analyses.
DISCUSSION
S. cerevisiae is the most widely used unicellular eukaryotic organism in biological research. Mass spectrometry-based proteomics has been used to identify a comprehensive proteome of S. cerevisiae (22). Yeast has also been used as a model organism in several MS-based global phosphoproteomic studies (78, 79).
Lysine acetylation has emerged as an important regulatory posttranslational modification. It is an evolutionary conserved modification that is found in all living organisms from bacteria to humans, suggesting that its regulatory roles may be conserved during evolution. The functional roles of lysine acetylation in histones have been extensively studied for over four decades, but until recently, deciphering the extent of this modification in nonhistone proteins remained challenging. In this study we use high resolution, high accuracy mass spectrometry-based proteomics to investigate the regulatory scope of acetylation in budding yeast. Identification of thousands of acetylation sites in this single cellular organism suggests that acetylation is at least as prevalent in yeast as in metazoans (26, 28). Our data shows that acetylation targets proteins involved in wide-ranging cellular functions, implying that this modification has broad regulatory roles. Considering the size of the yeast proteome, which is only about one-quarter of the human proteome, the number of acetylation sites identified in yeast is relatively large. Also, it appears that compared with phosphorylation, acetylation is relatively frequently observed in bacteria (28, 63–65, 80–82). However, the abundances of individual acetylation sites and functional implications of the relatively frequent occurrence of acetylation in prokaryotes and lower eukaryotes are presently unclear.
In accordance with a proposed evolutionary conserved regulatory role, we find that acetylated lysines are significantly more conserved across diverse species. It should be noted that a large fraction of conserved acetylation sites are present on proteins involved in vital biological processes that are conserved in essentially all living organisms, including metabolic enzymes, heat shock proteins, and ribosomes. A majority of enzymes involved in metabolic pathways are acetylated, often on multiple lysines. Similar observations have been made in metabolic enzymes from human (26, 27), Drosophila (28), and bacteria (64, 65) suggesting that acetylation may control activity or stability of these enzymes.
The use of HCD fragmentation in our analyses permitted us to explore the use of the previously described acetyl-lysine diagnostic ion (60) for confident identifications of acetylated peptides in mass spectrometry-based large-scale PTM analysis. The presence of the Im(acK) ion correlated well with the presence of acetylated lysines in the peptide sequences, however, a fraction of nonacetylated peptides also contained this ion in their fragment spectra. Our analyses show that in samples enriched for acetyl-lysine containing peptides, the presence of the Im(acK) ion in fragment spectra is a strong indicator of the presence of an acetylated lysine in the peptide sequence. However, a small fraction of fragment spectra of nonacetylated peptides also contain this ion, possibly derived from cofragmentation of acetylated peptides.
Quantitative analysis of rpd3Δ cells identified several acetylation sites, which showed increased abundance in rpd3 knockout cells, including several sites on histones and histone acetyltransferases. These results are in agreement with the previously known functions of Rpd3, and suggest that the strategy described here can be applied to identify putative substrates of other deacetylases in yeast. We identified a previously unknown Rpd3-regulated acetylation site on the SAGA complex subunit Sgf73. The functional significance of this modification remains to be investigated; however, previously published structure function data and our structural modeling analyses indicates that acetylation of this lysine may play a regulatory role by modulating the interaction between Sgf73 and Ubp8.
In summary, we report the first large-scale, high resolution MS-based survey of lysine acetylation in Saccharomyces cerevisiae. Our results show that acetylated lysines are evolutionary conserved, supporting an anciently conserved role of this modification. The provided dataset may serve as important starting point for assessing the functional relevance of acetylation sites in budding yeast.
Supplementary Material
Acknowledgments
We thank the members of the department of proteomics at CPR for their helpful discussions.
Footnotes
* This work is supported by the European Commission's 7th Framework Program grants Proteomics Research Infrastructure Maximizing knowledge EXchange and access (XS) (INFRASTRUCTURESF7-2010-262067/PRIME-XS), the European Research Council (ML), and the Lundbeck Foundation (R48-A4649). SAW is supported by a postdoctoral grant from Danish Council for Independent Research (FSS: 10-085134). The Center for Protein Research is funded by a generous grant from the Novo Nordisk Foundation.
 This article contains supplemental Figs. S1 to S4 and Tables S1 and S2.
This article contains supplemental Figs. S1 to S4 and Tables S1 and S2.
Data availability: All raw data associated with this manuscript will be provided on request.
1 The abbreviations used are:
- KAT
- lysine acetyl transferase
- KDAC
- lysine deacetylase
- HAT
- histone acetyl transferase
- HDAC
- histone deacetylase
- Lysine0
- L-lysine 12C6,14N2
- Lysine8
- L-lysine 13C6,15N2
- LTQ
- linear trap quadrupole
- HCD
- higher-energy C-trap dissociation
- SGD
- Saccharomyces genome database
- FDR
- false discovery rate
- eggnog
- evolutionary genealogy of genes: nonsupervised orthology group
- KOG
- eurkaryotic orthology group
- euNOG
- eukaryotic nonsupervised orthology group
- STRING
- Search Tool for the Retrieval of Interacting Genes/Proteins
- SILAC
- stable isotope labeling by amino acids in cell culture
- GOBP
- GO biological process
- KEGG
- Kyoto encyclopedia of genes and genomes
- MD
- molecular dynamics.
REFERENCES
- 1. Yang X. J., Seto E. (2007) HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310–5318 [DOI] [PubMed] [Google Scholar]
- 2. Kurdistani S. K., Grunstein M. (2003) Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4, 276–284 [DOI] [PubMed] [Google Scholar]
- 3. Yang X. J., Seto E. (2008) Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V., Jr., Alt F. W., Kahn C. R., Verdin E. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shahbazian M. D., Grunstein M. (2007) Functions of site-specific histone acetylation and deacetylation. Ann. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 6. Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U. S. A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guan K. L., Xiong Y. (2010) Regulation of intermediary metabolism by protein acetylation. Trends Biochem. Sci. 36, 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shimazu T., Hirschey M. D., Huang J. Y., Ho L. T., Verdin E. (2010) Acetate metabolism and aging: an emerging connection. Mech. Ageing Develop. 131, 511–516 [DOI] [PubMed] [Google Scholar]
- 9. Albaugh B. N., Arnold K. M., Denu J. M. (2011) KAT(ching) metabolism by the tail: insight into the links between lysine acetyltransferases and metabolism. Chembiochem. 12, 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guarente L. (2011) The logic linking protein acetylation and metabolism. Cell Metabol. 14, 151–153 [DOI] [PubMed] [Google Scholar]
- 11. Close P., Creppe C., Gillard M., Ladang A., Chapelle J. P., Nguyen L., Chariot A. (2010) The emerging role of lysine acetylation of non-nuclear proteins. Cell Mol. Life Sci. 67, 1255–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J., Shi X., Li Y., Kim B. J., Jia J., Huang Z., Yang T., Fu X., Jung S. Y., Wang Y., Zhang P., Kim S. T., Pan X., Qin J. (2008) Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31, 143–151 [DOI] [PubMed] [Google Scholar]
- 13. Imai S., Guarente L. (2010) Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 31, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jensen O. N. (2006) Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 7, 391–403 [DOI] [PubMed] [Google Scholar]
- 15. Choudhary C., Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 16. Macek B., Mann M., Olsen J. V. (2009) Global and site-specific quantitative phosphoproteomics: principles and applications. Ann. Rev. Pharmacol. Toxicol. 49, 199–221 [DOI] [PubMed] [Google Scholar]
- 17. Mischerikow N., Heck A. J. (2011) Targeted large-scale analysis of protein acetylation. Proteomics 11, 571–589 [DOI] [PubMed] [Google Scholar]
- 18. Beli P., Lukashchuk N., Wagner S. A., Weinert B. T., Olsen J. V., Baskcomb L., Mann M., Jackson S. P., Choudhary C. (2012) Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 46, 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., Galibert F., Hoheisel J. D., Jacq C., Johnston M., Louis E. J., Mewes H. W., Murakami Y., Philippsen P., Tettelin H., Oliver S. G. (1996) Life with 6000 genes. Science 274, 546, 563–567 [DOI] [PubMed] [Google Scholar]
- 20. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pal C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) The genetic landscape of a cell. Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boone C., Bussey H., Andrews B. J. (2007) Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8, 437–449 [DOI] [PubMed] [Google Scholar]
- 22. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 23. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Hofert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 24. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrin-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 25. Lee K. K., Workman J. L. (2007) Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 26. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 27. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weinert B. T., Wagner S. A., Horn H., Henriksen P., Liu W. R., Olsen J. V., Jensen L. J., Choudhary C. (2011) Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci. Signaling 4, ra48. [DOI] [PubMed] [Google Scholar]
- 29. Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Veronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- 30. Hubner N. C., Ren S., Mann M. (2008) Peptide separation with immobilized pI strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics 8, 4862–4872 [DOI] [PubMed] [Google Scholar]
- 31. Horth P., Miller C. A., Preckel T., Wenz C. (2006) Efficient fractionation and improved protein identification by peptide OFFGEL electrophoresis. Mol. Cell. Proteomics 5, 1968–1974 [DOI] [PubMed] [Google Scholar]
- 32. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 33. Olsen J. V., Schwartz J. C., Griep-Raming J., Nielsen M. L., Damoc E., Denisov E., Lange O., Remes P., Taylor D., Splendore M., Wouters E. R., Senko M., Makarov A., Mann M., Horning S. (2009) A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol. Cell. Proteomics 8, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelstrup C. D., Young C., Lavallee R., Nielsen M. L., Olsen J. V. (2012) Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole Orbitrap mass spectrometer. J. Proteome Res. 11, 3487–3497 [DOI] [PubMed] [Google Scholar]
- 35. Michalski A., Damoc E., Lange O., Denisov E., Nolting D., Müller M., Viner R., Schwartz J., Remes P., Belford M., Dunyach J. J., Cox J., Horning S., Mann M., Makarov A. (2012) Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol. Cell. Proteomics 11, 10.1074/mcp.O111.013698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 37. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 38. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 39. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protocols 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 40. Engel S. R., Balakrishnan R., Binkley G., Christie K. R., Costanzo M. C., Dwight S. S., Fisk D. G., Hirschman J. E., Hitz B. C., Hong E. L., Krieger C. J., Livstone M. S., Miyasato S. R., Nash R., Oughtred R., Park J., Skrzypek M. S., Weng S., Wong E. D., Dolinski K., Botstein D., Cherry J. M. (2010) Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 38, D433–D436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 42. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 43. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 44. Muller J., Szklarczyk D., Julien P., Letunic I., Roth A., Kuhn M., Powell S., von Mering C., Doerks T., Jensen L. J., Bork P. (2010) eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 38, D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 46. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protocols 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Köhler A., Zimmerman E., Schneider M., Hurt E., Zheng N. (2010) Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 50. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) Gromacs 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 51. Scott W. R. P., Hunenberger P. H., Tironi I. G., Mark A. E., Billeter S. R., Fennen J., Torda A. E., Huber T., Krüger P., van Gunsteren W. F. (1999) The GROMOS Biomolecular Simulation Program Package. J. Phys. Chem. A, 3596–3607 [Google Scholar]
- 52. Bussi G., Donadio D., Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 1–8 [DOI] [PubMed] [Google Scholar]
- 53. Parrinello M., Rahman A. (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 [Google Scholar]
- 54. Darden T., York D., Pedersen L. (1993) Particle mesh Ewald: an N log (N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 [Google Scholar]
- 55. Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M. (1997) LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 [Google Scholar]
- 56. Miyamoto S., Kollman P. A. (1992) Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 [Google Scholar]
- 57. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 58. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 59. Kim J. Y., Kim K. W., Kwon H. J., Lee D. W., Yoo J. S. (2002) Probing lysine acetylation with a modification-specific marker ion using high-performance liquid chromatography/electrospray-mass spectrometry with collision-induced dissociation. Anal. Chem. 74, 5443–5449 [DOI] [PubMed] [Google Scholar]
- 60. Trelle M. B., Jensen O. N. (2008) Utility of immonium ions for assignment of epsilon-N-acetyllysine-containing peptides by tandem mass spectrometry. Anal. Chem. 80, 3422–3430 [DOI] [PubMed] [Google Scholar]
- 61. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 62. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 63. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y., Ning Z. B., Zeng R., Xiong Y., Guan K. L., Zhao S., Zhao G. P. (2010) Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu B. J., Kim J. A., Moon J. H., Ryu S. E., Pan J. G. (2008) The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 18, 1529–1536 [PubMed] [Google Scholar]
- 65. Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., Zhao Y. (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. (2010) KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38, D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 [DOI] [PubMed] [Google Scholar]
- 68. Suka N., Suka Y., Carmen A. A., Wu J., Grunstein M. (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8, 473–479 [DOI] [PubMed] [Google Scholar]
- 69. Yuan H., Rossetto D., Mellert H., Dang W., Srinivasan M., Johnson J., Hodawadekar S., Ding E. C., Speicher K., Abshiru N., Perry R., Wu J., Yang C., Zheng Y. G., Speicher D. W., Thibault P., Verreault A., Johnson F. B., Berger S. L., Sternglanz R., McMahon S. B., Cote J., Marmorstein R. (2011) MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 31, 58–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rolef Ben-Shahar T., Heeger S., Lehane C., East P., Flynn H., Skehel M., Uhlmann F. (2008) Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321, 563–566 [DOI] [PubMed] [Google Scholar]
- 71. Unal E., Heidinger-Pauli J. M., Kim W., Guacci V., Onn I., Gygi S. P., Koshland D. E. (2008) A molecular determinant for the establishment of sister chromatid cohesion. Science 321, 566–569 [DOI] [PubMed] [Google Scholar]
- 72. Yang X. J., Seto E. (2008) The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rundlett S. E., Carmen A. A., Kobayashi R., Bavykin S., Turner B. M., Grunstein M. (1996) HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. U. S. A. 93, 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kadosh D., Struhl K. (1997) Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89, 365–371 [DOI] [PubMed] [Google Scholar]
- 75. Rundlett S. E., Carmen A. A., Suka N., Turner B. M., Grunstein M. (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392, 831–835 [DOI] [PubMed] [Google Scholar]
- 76. Samara N. L., Datta A. B., Berndsen C. E., Zhang X., Yao T., Cohen R. E., Wolberger C. (2010) Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328, 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Köhler A., Schneider M., Cabal G. G., Nehrbass U., Hurt E. (2008) Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nature Cell Biol. 10, 707–715 [DOI] [PubMed] [Google Scholar]
- 78. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 79. Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P. G., Gerrits B., Picotti P., Lam H., Vitek O., Brusniak M. Y., Roschitzki B., Zhang C., Shokat K. M., Schlapbach R., Colman-Lerner A., Nolan G. P., Nesvizhskii A. I., Peter M., Loewith R., von Mering C., Aebersold R. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signaling 3, rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 81. Soufi B., Gnad F., Jensen P. R., Petranovic D., Mann M., Mijakovic I., Macek B. (2008) The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics 8, 3486–3493 [DOI] [PubMed] [Google Scholar]
- 82. van Noort V., Seebacher J., Bader S., Mohammed S., Vonkova I., Betts M. J., Kuhner S., Kumar R., Maier T., O'Flaherty M., Rybin V., Schmeisky A., Yus E., Stülke J., Serrano L., Russell R. B., Heck A. J., Bork P., Gavin A. C. (2012) Cross-talk between phosphorylation and lysine acetylation in a genome-reduced bacterium. Mol. Syst. Biol. 8, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.