Fig. 1.
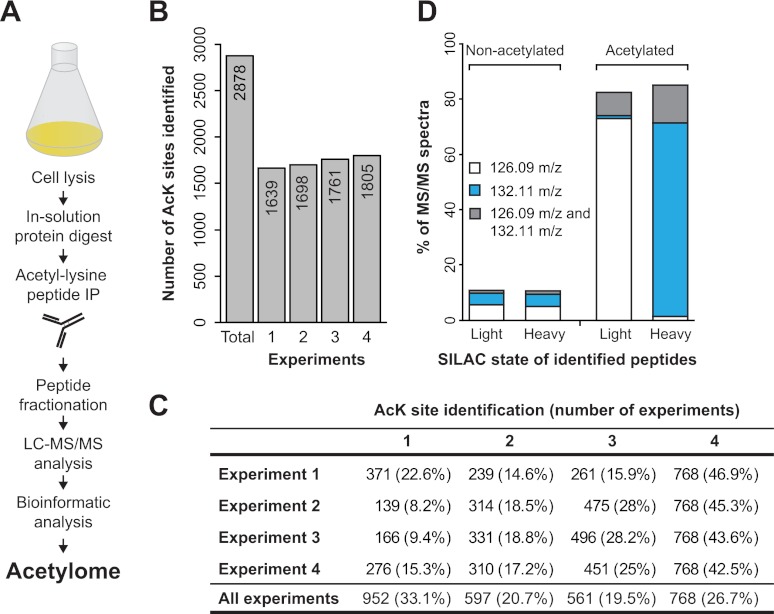
Proteome-wide identification of lysine acetylation sites in S. cerevisiae. A, Schematic representation of the workflow used for the identification of yeast acetylation sites. Yeast cells were harvested and mechanically lysed, and proteins were digested into peptides and acetylated peptides were enriched using anti-acetyllysine antibodies. The enriched peptides were further fractionated by isoelectric focusing and analyzed by mass spectrometry. B, The graph shows the number of acetylation sites identified in each of four experiments, as well as total number of acetylation sites identified in all four experiments. The number of identified acetylation sites is indicated within each bar. C, The table shows the number of acetylation sites identified in each experiment and the percent of acetylation sites identified in one or more experiments. For example, for any of four experiments 77–92% of sites were identified in more than one experiment. D, The presence of the acetyllysine diagnostic ion in MS fragment scans of acetylated peptides and nonacetylated peptides. The mass of the diagnostic ion (126.09 m/z or 132.11 m/z) is closely associated with the SILAC state of the lysine from corresponding fragment scans.