Fig. 3.
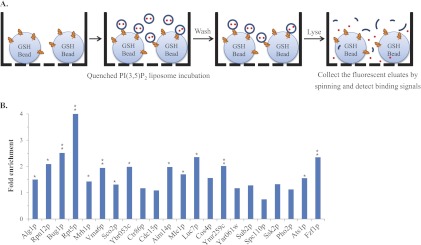
The bead-based affinity assays confirmed the PI(3,5)P2-protein interactions revealed by the NQF liposomes. A, A schematic representation of the bead-based affinity assay. The proteins of interest were purified by the GST-glutathione (GSH) bead affinity purification procedure (9). Prior to elution, fabricated PI(3,5)P2 fluorescent liposomes were added into each well of a 96-filter plate and allowed to interact with the purified target proteins. After the complete removal of unbound liposomes, the bound liposomes were lysed with 30 mm n-OG, and the fluorescent eluates were collected in a 96-well black plate to detect each binding signal. B, Validation of identified PI(3,5)P2-protein interactions. We chose 29 PI(3,5)P2-BPs to conduct the bead-based affinity assays, and each protein sample was conducted in triplicate. For normalization, the binding signals of the proteins were divided by the signal of the glutathione beads without proteins to generate the fold enrichment. Among the 29 proteins, seven were not detectable on an SDS-PAGE gel stained with Coomassie brilliant blue (data not shown). Therefore, 14 (64%) of the 22 proteins showed more than a 1.3-fold enrichment with statistical significance (* p < 0.05, ** p < 0.005; n = 3).
