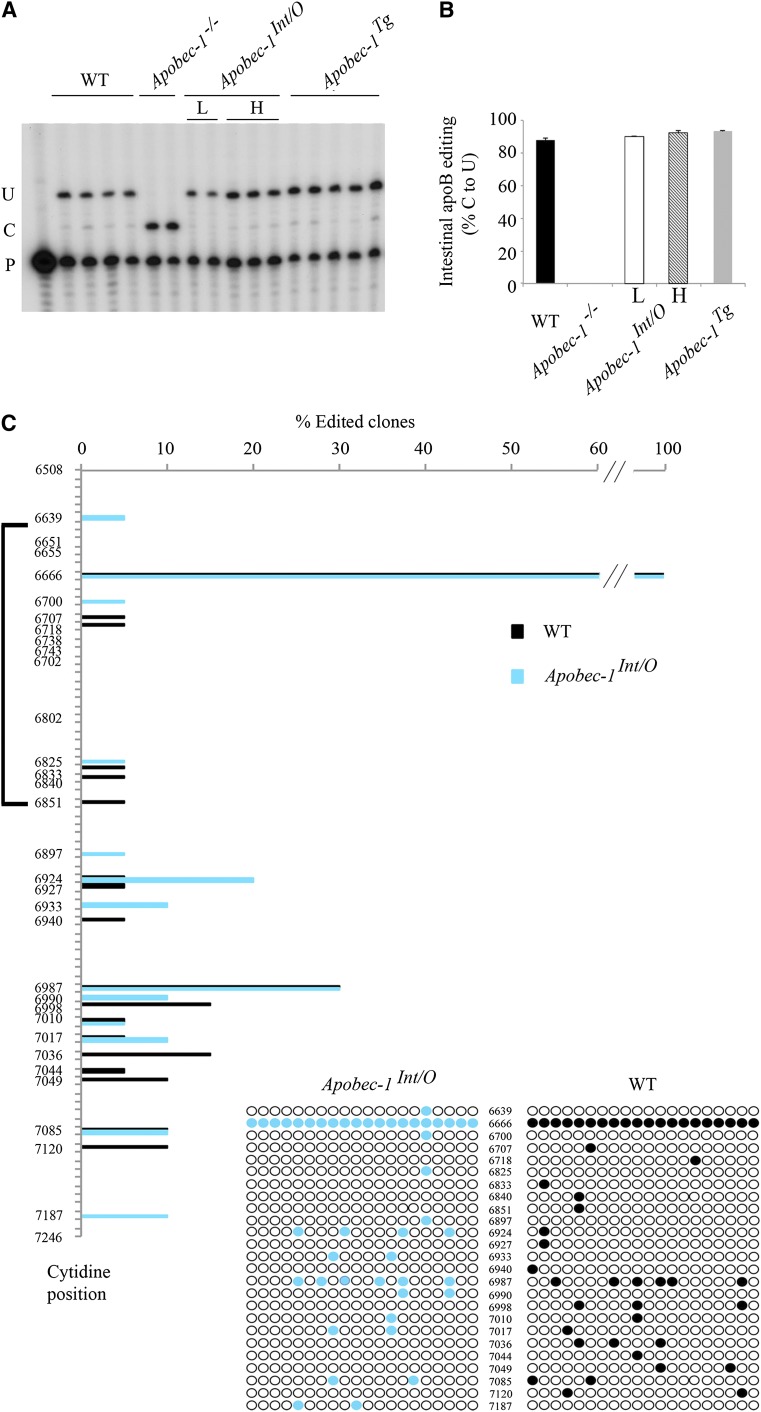
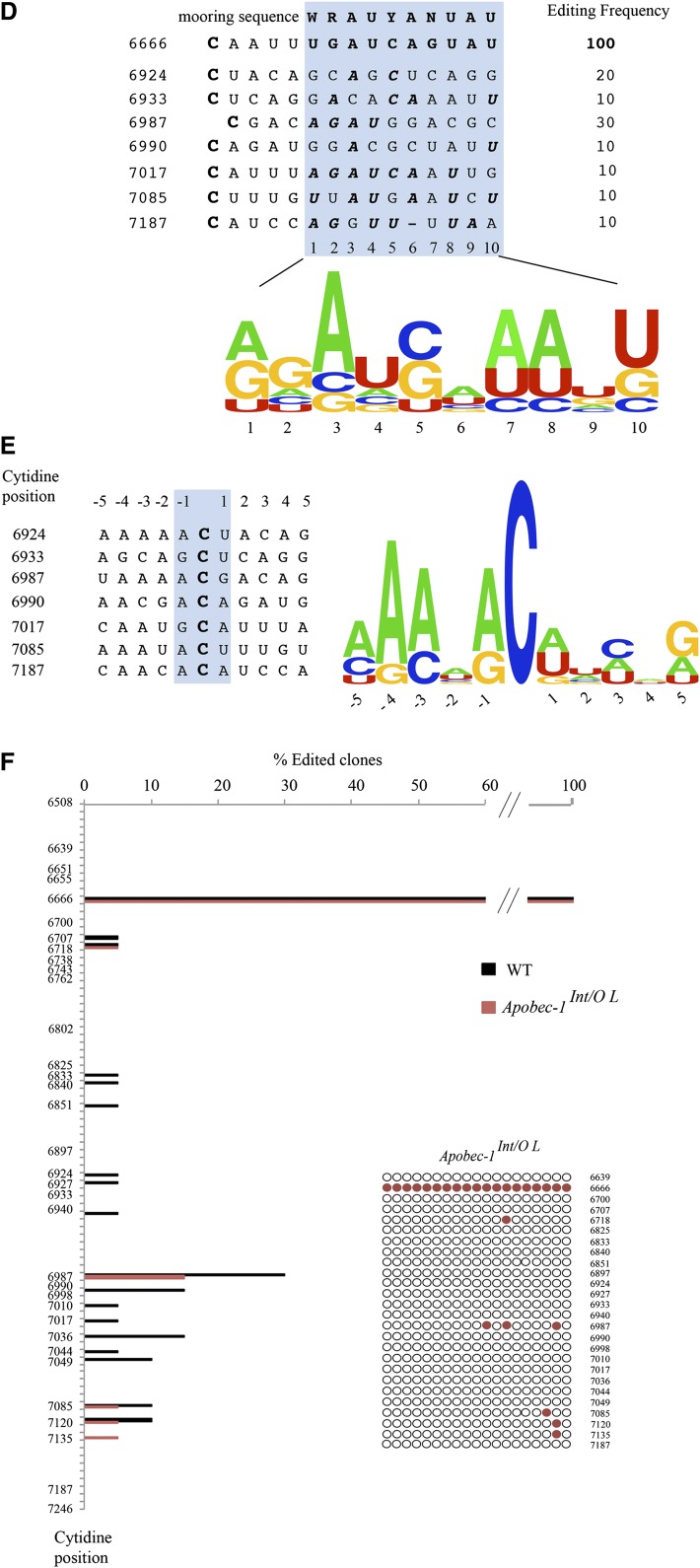
Fig. 2.
Transgenic rescue of apobec-1 in the intestine of Apobec-1−/− mice restores ApoB C-to-U RNA editing. A: Endogenous apoB mRNA editing was determined by primer extension. The relative mobility of the unedited (C) and edited (U) products is indicated on the left. B: Bar graph representation of percentage C-to-U editing for each group as mean ± SE. (n = 3–5). C, D: Hyperediting of apoB mRNA analyzed by DNA sequencing. A 738 bp fragment (6508–7246 nt) of apoB mRNA overlapping the C6666 canonical editing site was amplified by RT-PCR subcloned into pPCR-Script vector and sequenced. Twenty clones per genotype (from two mice per genotype) were analyzed. Targeted cytidines identified in Apobec-1Int/O clones are indicated by blue circles, aligned with the nucleotide position. Edited cytidines identified in WT clones are indicated with black circles, aligned with the nucleotide position. The bracket on the left side of the vertical panel indicates the region of apoB mRNA found by Innerarity and colleagues to be promiscuously edited in hepatic apobec-1 transgenic mice (20). D: Alignment of a 14 nt sequence downstream of C-to-U editing sites with ≥ 10% editing identified in RNA from Apobec-1Int/Oh mice [see (C)]. The sequences were compared with the apoB RNA canonical mooring sequence shown on the top. W, A/U; R, A/G; Y, U/C. The edited C is indicated in bold font at the beginning of each sequence. The nucleotides matching the mooring sequence are indicated in bold italic letters. Below is shown the frequency plot of nucleotides in the 10 nt motif containing matching residue aligned with the consensus mooring sequence (blue box). E: Alignment and frequency plot of nearest neighbor nucleotides flanking each C-to-U RNA editing site. Positions are indicated relative to the targeted cytidine. Frequency plots were created using Weblogo software (Berkeley, CA). F: Hyperediting of intestinal apoB RNA isolated from Apobec-1Int/O low-expressor mice and analyzed by sequencing as in (C). Edited cytidines identified in 20 independent clones are indicated by red circles.