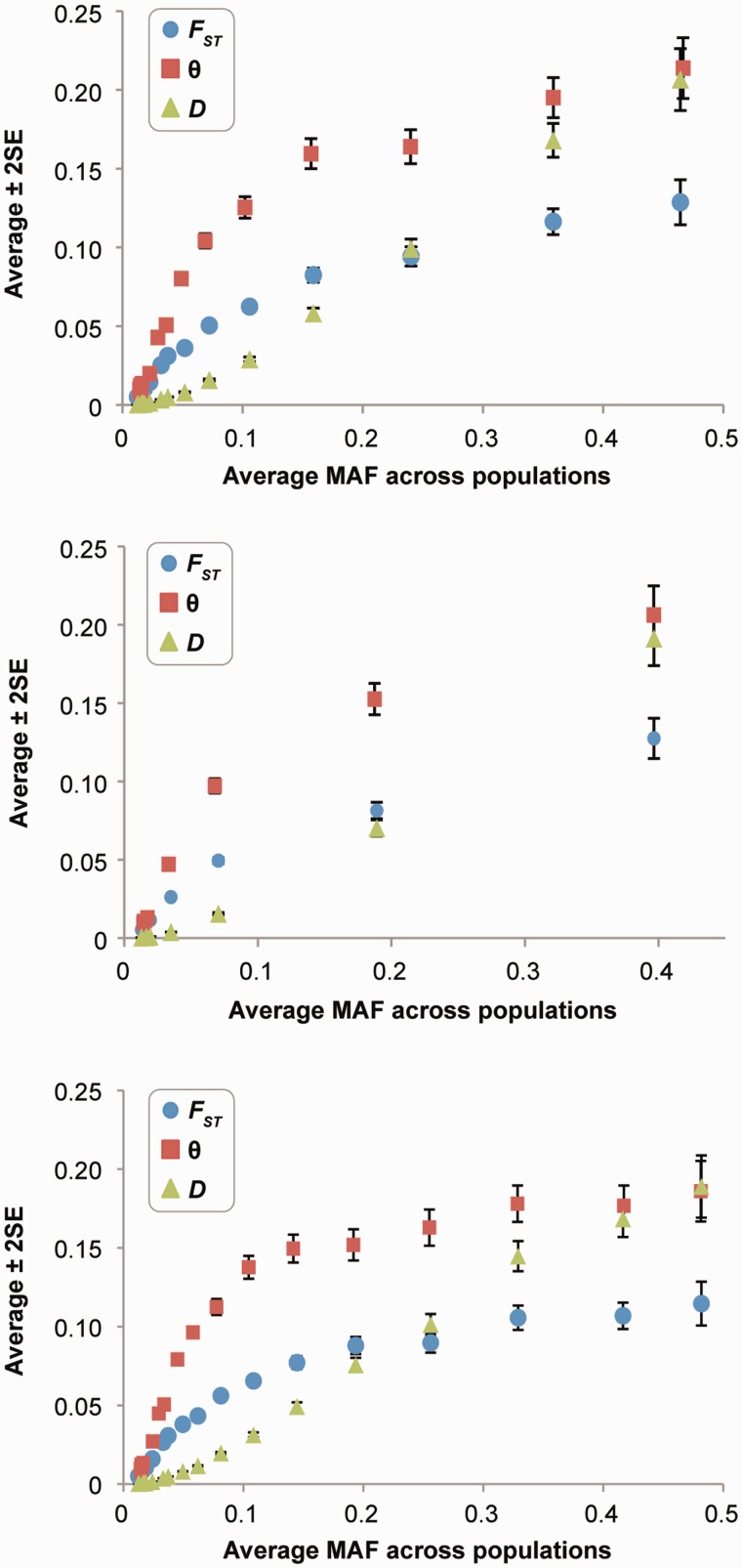
Fig. 3.
The relationship between minor allele frequency (MAF) and population differentiation estimates for nonsynonymous SNPs (nSNPs). Each point shows the average estimate (± two standard errors) in nonoverlapping sliding windows sorted by MAF of 1,000 SNPs, except for positions with the highest MAF. For nSNPs, (A) shows the relationship from all loci. The correlation coefficients of the raw data (sliding window) are 0.60 (0.95), 0.58 (0.91), and 0.71 (1.00) for FST, θ, and D, respectively. They are all significant at P < 10−15. In (B), the relationship in (A) is restricted to including one nSNP from each protein to avoid linkage effects. In (C), the relationship is shown for synonymous SNPs. In this panel, the correlation coefficients between MAF and FST, θ, and D are 0.53 (0.92), 0.52 (0.86), and 0.67 (1.00), respectively, which are all significant at P < 10−15.