Figure 11.
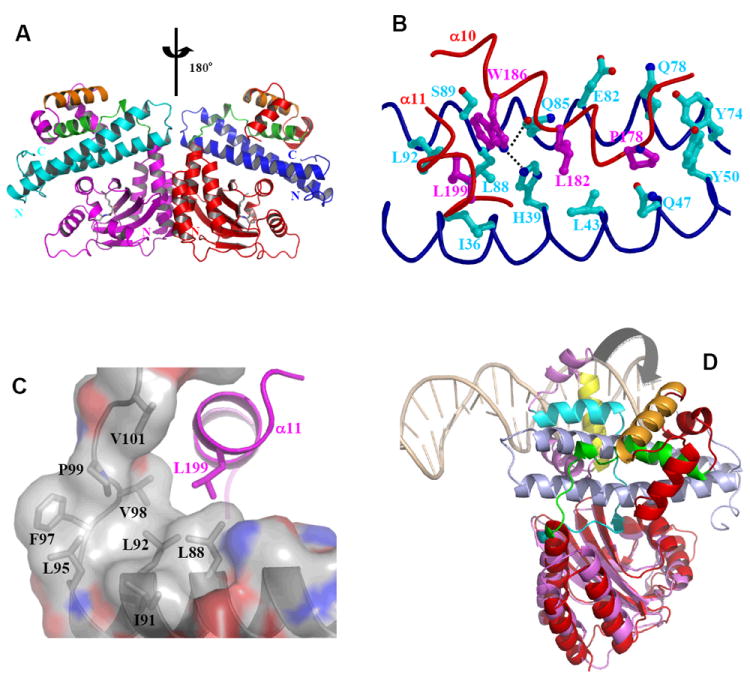
Structure and function of anti-activation by TraM. A) Model of symmetric (TraRNGR-TraMNGR)2 in solution. The model was generated by applying the C2 rotational symmetry of the NTDs to the closed form of dimeric TraRNGR-TraMNGR. The repositioned TraRNGR-TraMNGR pair is colored in red and blue, and the original pair is in magenta and cyan. The main TraM-binding site, α10 and the linker, is in green, and the DNA binding helix, α12, is in orange. The bound 3-oxo-C8-HSL is shown in ball-and-stick representation. B) The interaction between TraRNGR α10 with TraMNGR (PDB ID: 2Q0O). TraMNGR residues (cyan) that are in contact with Pro178, Leu182 and Trp186 (magenta) of TraRNGR α10 are shown. Leu199 from α11 is also shown. The dotted lines denote hydrogen bonds between TraRNGR Trp186 and TraMNGR His39 and Gln85. C) The interaction of TraRNGR L199 with the hydrophobic C-terminal tail of TraMNGR. Surface is colored according to the underlying atoms: red for oxygen, blue for nitrogen, and gray for carbon. Side chains of the hydrophobic TraMNGR residues (gray), along with that of L199 (magenta), are shown. D) Structural comparison of TraRAt-DNA with TraRNGR-TraMNGR. The NTDs of the both structures are superimposed, but for clarity only one monomer of TraRNGR (in red) and TraRAt (in purple, the extended form) from each structure is shown. The large domain movement is highlighted with an arrow. DNA is displayed as a double coil and is light orange, and the DNA-binding helix α12 is yellow in TraRAt-DNA complex and orange in TraRNGR-TraMNGR complex. TraMNGR is light blue, and the TraM-binding site (α10 and the linker) is cyan in TraRAt-DNA and green in TraRNGR-TraMNGR.
