Figure 8.
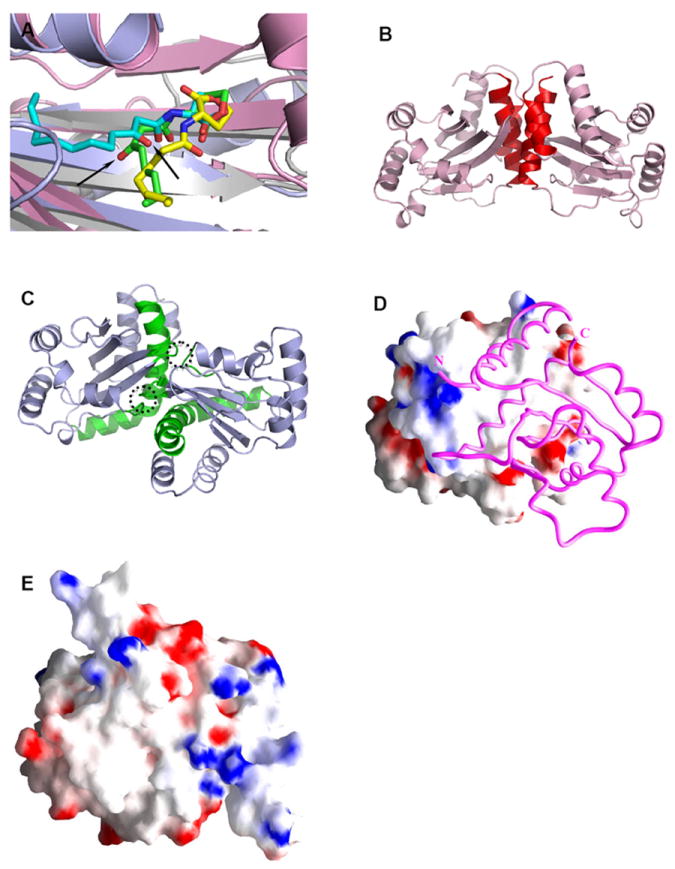
Structural comparison of the LuxR-type proteins. A) Conformations of AHLs in the AHL-binding pockets: 3-oxo-C8-HSL (green) in TraRAt (pink, PDB ID: 1L3L); 3-oxo-C12-HSL (cyan) in LasR (light blue, PDB ID: 2UV0); C8-HSL (yellow) in SdiA (gray, PDB ID: 2AVX). For clarity, structural elements (helices and loops) that are located on the top of the bound AHLs are removed. The different orientations of the 3-oxo group in 3-oxo-C8-HSL and 3-oxo-C12-HSL that lead to drastic conformations of the acyl chains are highlighted with arrows. B) and C) Distinct dimeric assembly of TraR and LasR shown in the same view. Structure of dimeric TraRAt NTD (pink) is overlaid with that of LasR NTD (light blue) by superimposing one (left) NTD of each dimer. The main helices that are located in the TraRAt dimer interface are largely parallel and highlighted in red, and those in LasR are almost perpendicular and in green. The dimer contacts provided by loops of α4-β3 and β4-β5 in LasR are highlighted in dotted circles. D) and E) Surface electrostatic potentials of TraRAt and SdiA, respectively. Surface of one TraRAt NTD is shown, the other NTD is shown as a Cα trace in magenta. The view of SdiA corresponds to that with the surface presentation of TraRAt monomer. Surface with positive potentials is colored in blue, and with negative potentials in red.
