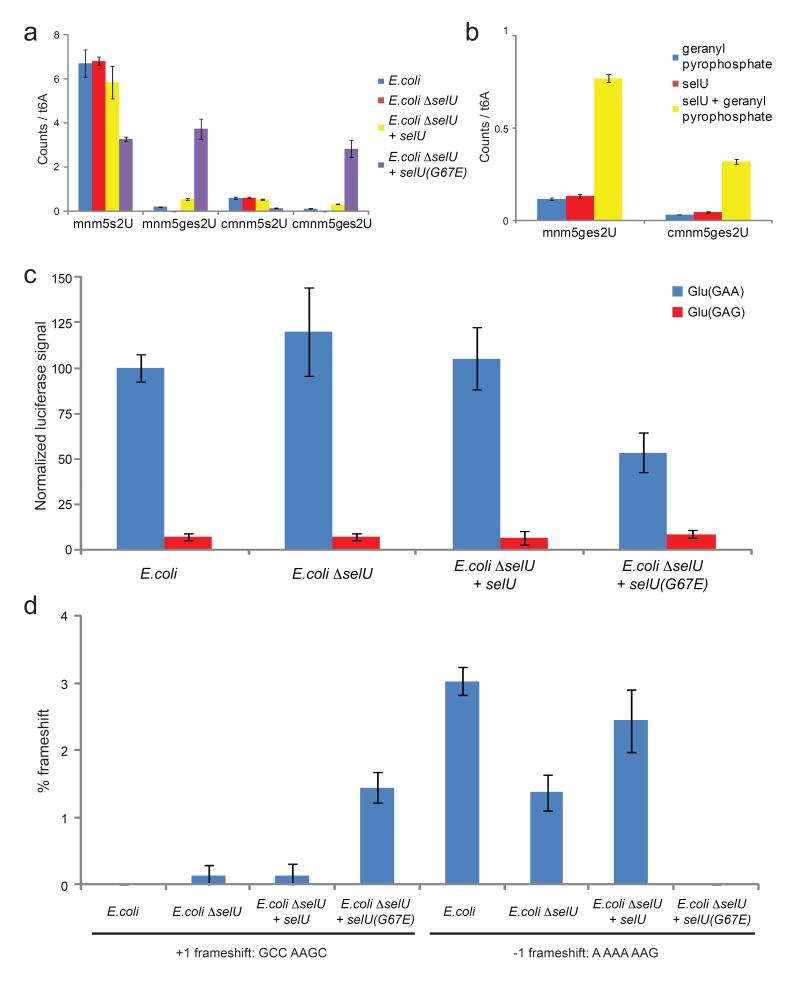
Figure 5.
Biological abundance and properties of geranylated RNA. (a) E. coli selU mutations affect geranylated nucleotide levels. No geranylated nucleotides were detected in E. coli cells lacking selU (ΔselU). Complementation with wild-type selU restores geranylation. Complementation with mutant selU(G67E) results in high levels of geranylation. Nucleoside levels are shown as relative ratios of MS counts of the individual nucleosides versus t6A. Error bars represent the standard deviation of three independent biological replicates. (b) SelU geranylates mnm5s2U and cmnm5s2U on tRNA in vitro in a geranyl pyrophosphate-dependant manner. Nucleoside levels are shown as relative ratios of MS counts of the individual nucleosides versus t6A. Error bars represent the standard deviation of three analytical replicates. (c) Luciferase reporter assay of glutamate codon translation efficiency for GAA and GAG reveals a strong bias favoring GAA translation under low geranylation conditions and a significantly lower preference under high geranylation conditions due to reduced efficiency of GAA decoding. Error bars represent the standard deviation of three independent biological replicates. (d) +1 and −1 frameshift efficiency at the sequences GCC AAGC and A AAA AAG inserted between GST and MBP reading frames. The Y-axis indicates the amount of GST-MBP fusion protein detected by western blot as a percentage of total GST-containing proteins produced. Error bars reflect the range of two technical replicates.