Abstract
The function and regulation of MDM2 as a component of a p53-dependent negative feedback loop has formed a core paradigm in the p53 field. This concept, now 20 years old, has been solidified by fields of protein science, transgenic technology, and drug discovery in human cancer. However, it has been noted that a simple negative feedback loop between p53 and MDM2 lacks an intrinsic “activating” step that counteracts this inhibition and permits oscillation of the feedback to occur as p53 is switched on and off. More recent work has identified a solution to the missing piece of the picture that counters the negative feedback loop, which is MDM2 itself. Under conditions of genotoxic stress, MDM2 helps to activate p53 by increasing its rate of protein synthesis. This simple observation makes certain aspects of the p53 response more comprehensible such as why MDM2 is upregulated by p53 early on following DNA damage and how phosphorylation of MDM2 at the C-terminal Ser395 by ATM translates into p53 activation. The latter acts by inducing allosteric changes in the RING domain of MDM2 that expose its RNA binding pocket, support p53 synthesis, and suppress its degradation. This allosteric nature of MDM2 in the C-terminus mirrors the allosteric effects of the binding of small molecules to the p53 interacting pocket at the N-terminus of MDM2, which opens the core domain of MDM2 to central domains of p53, which controls p53 ubiquitination. Thus, the highly allosteric nature of MDM2 provides the basis for dynamic protein-protein interactions and protein-RNA interactions through which MDM2’s activity is regulated in p53 protein destruction or in p53 protein synthesis. We discuss these mechanisms and how this information can be exploited for drug development programs aimed at activating p53 via targeting MDM2.
Keywords: Mdm2, p53, allosteric regulation
The p53-MDM2 feedback loop is proving to be important not only for cancer control but also for tissue injury associated with aging. These diverse biological outcomes depend on the inputs whereby p53 can be activated and affirm that relatively modest changes in the levels of p53 have important consequences for cell biology and animal physiology, underlining the importance of controlling p53 expression levels under normal conditions as well as under conditions of cellular stress. For example, pharmacological activation of p53 in murine transgenes using the C-terminal peptide domain induces a pro-aging phenotype such as reduced longevity, osteoporosis, generalized organ atrophy, and a diminished stress tolerance osteoporosis.1 Enhanced pro-aging is also seen from p53-induced senescence in murine skin after reducing MDM2 gene dosage.2 A second transgenic study showing that enhanced p53 function promotes aging used another truncated form of p53 with mutations in the MDM2-binding domain.3 An additional transgenic model displaying a pro-aging phenotype had a BRCA1 mutation that constitutively activates p53 via the enhanced endogenous DNA damage signals.4 Furthermore, chronic adrenaline stress through β-androgenic receptors mediates MDM2 suppression and activates p53 to accelerate DNA damaging and aging in mice.5 These data together suggest that “activation” of p53 can be linked to tissue injury linked to aging. This might seem in contrast to the esteemed guardian of the genome function of p53; indeed, murine transgenes in which p53 gene dosage is elevated (“triploid” p53) are cancer resistant and, interestingly, show no signs of rapid aging seen by the “pharmacological” activation of p53.6 Furthermore, the creation of a transgenic animal with elevated ARF and elevated p53 gene dosage is not only cancer resistant but lives longer,7 thus showing that p53 can either induce tissue aging or be involved in reducing tissue aging depending on the “input” signal. This highlights the interest in the scientific community for understanding how the specific activity of p53 is regulated by MDM2 and how this might lead to quite diverse biological outcomes.
There are multiple levels at which MDM2 and p53 can be regulated, from their birth (mRNA production and protein synthesis), to their life (protein trafficking to quite distinct compartments where they interact with hundreds of distinct binding proteins), to their death (protein destruction). The spatiotemporal regulation of the hundreds of protein-protein interactions within a hub8 remains undefined; indeed, how even 2 or 3 different hubs such as the “early” identified ATM-p539 are trafficked or the latest ATG7-p53 hub10 remains undefined. Understanding how the different p53 protein-protein interactions are integrated with p53 modifications and which protein-protein interactions depend on one another or are mutually exclusive forms a daunting task. What is clear, however, is that the levels of p53 mRNA generally do not change during p53 activation, and the important regulatory mechanisms of p53 expression levels are on the p53 protein turnover and its rate and synthesis. This allows for the cells to rapidly control the levels of p53 and reduces the number of steps required to activate p53 that can be affected by cellular DNA damage or physiological inputs such as immune system signaling through virus infection. In addition to change in the levels of expression, p53 is also subject to a vast number of different modifications that regulate its activity as a transcription factor, allowing the cells to induce cellular responses that best match the type of cellular damage that leads to p53 activation. Simplified, p53 activation is a 2-step process in which its expression is first stabilized whereupon its activity can be modified. The latter has been extensively reviewed elsewhere, and here we instead focus on some more recent advances in our understanding of the complex but elegant mechanism of how MDM2’s activities toward p53 expression are regulated by conformational changes in MDM2, and we discuss how this information can open alternative drug development strategies aimed at controlling p53 levels and activity.
How MDM2 Inhibits p53: A Negative Feedback Loop Operating at the Level of Protein Degradation
MDM2 has emerged as the key regulator of p53 expression based on mice and in cellulo models and from clinical samples showing it is amplified in several human cancers, most notably sarcomas. Mice lacking MDM2 die early during embryogenesis in a p53-dependent manner, and the MDM2-p53 interaction is conserved during evolution, suggesting an interesting coevolutionary process that might not only control p53 but equally well regulate MDM2 activity.11 MDM2 is best characterized for its capacity to promote its degradation, but MDM2 can also suppress p53 activity by direct interference with p53’s N-terminal transactivation domain. This aspect is somewhat less studied and could be restricted to affect some, but not all, of p53’s transactivity as p53 harbors at least 2 domains that can affect gene regulation, one of which includes the MDM2 binding to a motif in the transactivation domain of p53.12,13
Mechanistic studies have shown key events in how MDM2 can promote p53 ubiquitination in vitro, and the general consensus is that MDM2 also directly promotes p53 ubiquitination in vivo. It is, however, not yet known if MDM2 can cooperate with other ubiquitin ligases known to interact with p53, such as TRIM25 or PirH2, or if each ligase acts independently. Whether MDM2 promotes p53 polyubiquitination or monoubiquitination or, indeed, if these are options can be regulated or whether they have distinct signaling functions is also less clear. A fact that makes this trickier is that monoubiquitination of p53 can take place on several lysine residues in the p53 C-terminus as defined by mutagenesis studies, making it sometimes difficult to distinguish one event from the other.14 In addition, more recent research using mass spectrometry has also shown that the central domain of p53, not the C-terminus, contains the dominant acceptor sites for ubiquitination by MDM2 or PirH2 E3 ubiquitin ligases,15 and thus the actual site of ubiquitination in vivo remains to be defined accurately and might depend on the input signal and protein complexes assembled to drive combinatorial modifications on p53.
Regardless of the actual sites of ubiquitination of p53, the initial step in MDM2-dependent ubiquitination of p53 is the binding of the conserved peptide motif in the N-terminus of p53 (named the BOX-I domain) to a hydrophobic pocket in the N-terminus of MDM2. This interaction has been thoroughly studied, and numerous molecules have been developed that can compete for this interphase in the hope of preventing MDM2-mediated suppression of p53 and thereby activate p53 in cancers that express high levels of MDM2 and wild-type p53.16 Interestingly, however, is that this was thought to be the sole interaction between p53 and MDM2 required to promote p53 ubiquitination. But in fact, using molecules such as the nutlins that mimic the p53 BOX-I binding to MDM2 has revealed that this interaction is the first of a series of dynamic, transient protein-protein interactions that lead up to p53 ubiquitination. p53 or its mimetics, which bind the hydrophobic pocket of MDM2, alter the conformation of MDM2 allosterically so that more central domains of MDM2 are exposed to the core domain of p53,17 and this second interphase is required for the C-terminal RING domains of MDM2 to promote the E2 interaction18 and the ubiquitination of p53. These results demonstrate an elegant example of how disordered domains in both proteins act together to generate a diversified and dynamic regulation of p53 stability. Hence, these observations show how domains throughout the 2 oligomeric proteins are involved in a click-clack series of events that start in the N-termini and finish by bringing multidomains from both proteins in correct positions to recruit the E2 and promote ubiquitination on selected lysine residues (Fig. 1). This concept is further underlined by the stabilizing pseudo-substrate motif (i.e., “lid”) near the N-terminal hydrophobic pocket of MDM2 that can regulate the extent of allosteric activation of MDM2 toward p53 as well as a C-terminal tail that can sit in the RING domain and stabilize RING domain oligomers, including hetero-oligomers, with its homologue and MDMX.19 This highlights the allosteric nature of MDM2 and how modifications or interactions in one domain of MDM2 can bring about intramolecular alterations in separate domains. Nevertheless, how the N- and C-termini of MDM2 in conjunction with p53 domains are assembled into an “ubiquitination machine” that can shape a scaffold that bridges the E2-ubiquitin conjugate, as well as whether any stereochemistry orchestrates recognition of this tetravalent substrate, remains undefined. This lack of knowledge, relative to the cullin-ubiquitin machine as an example,20 is mainly due to the difficulty in crystallizing full-length MDM2 with tetrameric p53 due to the large degrees of intrinsically disordered domains required for their function. Nevertheless, these multiple, allosteric, and dynamic protein-protein interactions between MDM2-p53 and MDM2-E2 are a challenge to understand, but at the same time, they provide novel opportunities for therapeutic intervention in human disease.
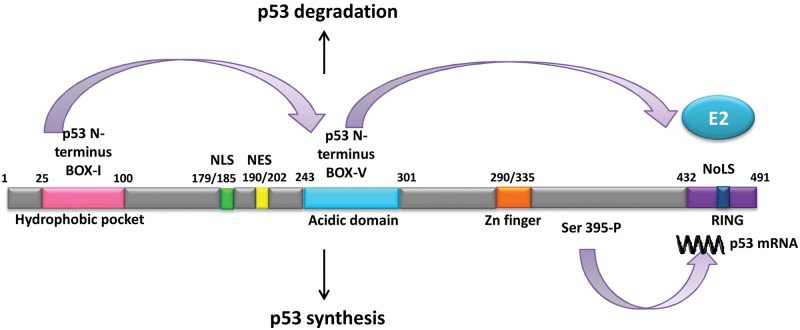
Figure 1.
(Upper part) Allosteric changes in Mdm2, mediated by ligand binding to the N-terminal hydrophobic pocket mimicking the BOX-I of p53, promote interaction between core domains of MDM2 and the conserved BOX-V domain of p53, which recruits the E2 ligase to the C-terminal RING domain and p53 ubiquitination and suppression of p53 activity. (Lower part) Phosphorylation of MDM2 at Ser 395 has the opposite effect on p53 expression as it opens up the RING domain for p53 mRNA binding, which suppresses MDM2 ligase activity and promotes p53 synthesis. Domains and localization sequences are indicated (localization sequences: NLS = nuclear; NES = nuclear export; NoLS = nucleolar).
How MDM2 Activates p53: A Positive Feedback in Protein Synthesis
It is intuitive that controlling the levels of p53 expression should involve both regulation of synthesis and degradation as both are equally important to reach a functional steady-state level. But it is, however, surprising to see how intimately linked these 2 events are and how a signaling pathway can tip the balance between synthesis and degradation to increase, or decrease, p53 expression levels. The BOX-1 of p53 is highly conserved also on a nucleotide level. The sequence of the p53 mRNA that encodes the BOX-1 and the MDM2-interacting amino acids also interacts directly with the C-terminal RING domain of MDM2 and promotes MDM2-mediated translation of the p53 mRNA.21 Thus, from the same genomic sequence of p53 has evolved 2 MDM2-interacting motifs—1 protein and 1 RNA—but, interestingly, with opposite functions toward p53 as the protein-protein interaction controls p53 turnover rate and the RNA-MDM2 interaction regulates, primarily, its rate of synthesis. The conserved nature of this sequence of p53 and the fact that it appears early during evolution together with MDM2 implies that the capacity to control the levels of p53 expression by coregulating synthesis and degradation has evolved together and that both functions might be equally important. An interesting aspect would be to know which, if so, of these interactions evolved first? Was the origin of this intimate relationship between MDM2 and p53 to promote or to suppress p53 expression?
p53 activation in response to genotoxic stress is critical for maintaining genomic integrity and for p53 tumor suppressor activity, and the first evidence that this involves an increase in p53 synthesis rate came from irradiation treatment of ML-1 cells in which the presence of cycloheximide prevented p53 accumulation.22 Supporting this notion, several groups using metabolic labeling with 35S-methionine and subsequent immunoprecipitation of p53 protein could show that newly synthesized p53 accumulates quickly in the cell following DNA damage caused by infrared radiation (IR),23-25 short ultraviolet (UVC) light irradiation,26 or etoposide.27 Thus, p53 accumulates in the cell mainly as a result of decreased p53 degradation by the E3 ubiquitin ligase MDM2 in response to genotoxic stress but also by increased p53 mRNA translation.
A more detailed model for how the DNA damage response controls p53 synthesis and degradation came from recent observations that the binding of MDM2 to the p53 mRNA is regulated and requires phosphorylation on MDM2 serine 395 by the ATM kinase22,28,29 (Fig. 2). But this site does not appear to be part of the direct interphase of the protein-RNA interaction as an MDM2 construct, which starts at 396 and thus does not include 395, still binds RNA with high affinity in a nonmodified fashion. Instead, it appears that phosphorylation on 395 opens up the RNA binding pocket of MDM2 (Fig. 1). Interestingly, small unspecific RNA oligonucleotides bind MDM2, but at the same time MDM2 binds larger mRNAs with high specificity. Together with the observation that a single silent point mutation in codon 22 (leucine) of the p53 mRNA is sufficient to severely weaken the interaction with MDM2, this implies that MDM2 has an RNA binding pocket with little specificity but that access to this pocket is highly specific and that RNAs with only the correct structure will be allowed access. This might reflect that the RNA interaction constitutes more than 1 site in the MDM2 RING domain and that the folding of the p53 mRNA plays an equally important role in MDM2-dependent synthesis of p53 and that this can be subject to regulation. In support of such a notion is the observation that the RNA sequence that binds to MDM2 includes a structure that has been shown to promote cap-independent translation, and such internal ribosome entry sites (IRESs) are known to constitute 3-dimensional structures to promote a regulated interaction with various factors of the translation machinery.30-32 It is likely that the MDM2-RNA interaction is not restricted to the p53 mRNA and includes other RNAs, and it was reported that MDM2 also binds the IRES of the XIAP mRNA.33
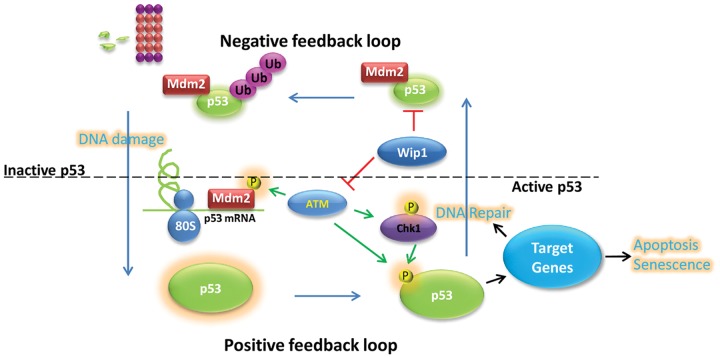
Figure 2.
The switch of MDM2 from a negative to a positive regulator of p53 is controlled by ATM kinase activity. Under normal conditions, p53 activity leads to an induction of MDM2 gene expression that targets p53 for polyubiquitination and degradation, keeping p53 activity low. Simplified, following DNA damage, ATM phosphorylates MDM2 at Ser395, which allows MDM2 to bind the p53 mRNA and promote p53 synthesis, switching the negative feedback loop to a positive. At the same time, modifications on p53 either direct or indirectly via ATM help to modify p53 activity. Depending on the nature and severity of the damage and cell type, the cell can either repair the damage or enter senescence or apoptosis.
Expression of the wild-type p53 protein from an mRNA with low affinity for MDM2 fails to stabilize p53 expression following treatment with DNA damage-inducing drugs. This is due to both a reduction in p53 synthesis and to a suppression of MDM2-mediated degradation of p53.21 Thus, the p53 mRNA interaction serves to switch MDM2 from a negative to a positive regulator of p53 via a double mechanism that includes an increase in p53 synthesis and the prevention of MDM2-dependent ubiquitination of p53. How the regulation of MDM2 E3 ligase activity is controlled by the p53 mRNA is not clear, but it is likely to mimic that previously observed for small nucleotides binding MDM2.
Within this switch from a negative to a positive regulator of p53 during stress lies also the localization of MDM2 to the nucleolus.28,34 Although the canonical roles of the nucleolus being rRNA transcription, pre-rRNA processing and nascent ribosome subunit assembly, it is evident that other nonribosomal maturation functions also take place.35,36 Both the p53 mRNA and nucleotide binding can promote MDM2 nucleolar localization, and it is likely that both ligands act on the same mechanism, and RNA interactions as signals for nucleolar targeting are also observed for other proteins such as NRF and nucleolin.37-39 The role of MDM2 nucleolar targeting might still be under debate, but these observations suggest that p53 mRNA bound to MDM2 can initiate an mRNP complex in or close to the nucleoli that will be transported to the cytoplasm for translation during DNA damage. However, nucleolar accumulation of MDM2 can also be mediated by the interaction with the p14Arf, which binds the central acidic domain of MDM2 and seems to have little effect on p53 synthesis, indicating that the accumulation of MDM2 in this compartment likely serves more than one function.40,41 The core MDM2 interactome appears to involve several ribosomal factors (L5, L11, L23, L26, S7, S3, 5S rRNA),42-44 which have an inhibitory interaction for ubiquitin ligase activity to stabilize p53, and it will be interesting to see how these interactions in conjunction with MDM2 nucleolar localization play a role in determining the rate of p53 of synthesis.
Perspectives
Recent years have seen the emergence of the MDM2-p53 interaction as highly dynamic and subject to regulation in order to control the outcome of p53 activity. The binding to ligands or the modifications by phosphorylation result in profound allosteric changes in MDM2 that span from one end of the protein to the other. The effect of these changes in structure has so far mainly been studied in terms of changes in p53 activity, but it is important to keep in mind that MDM2 also interacts with over 100 other proteins45 as to perhaps function as an oncoprotein in its own right. Some of these MDM2 binding proteins affect cell cycle control, including pRb and E2F, or transcription regulators such as p300 or transcription factors, or they regulate ageing via the β-adrenergic signaling axis as well as ribosomal biogenesis via interaction with ribosomal factors. All these interactions can together, or individually, contribute to tumor development and growth. Even though the regulation of these interactions is less studied as compared with p53, there is no reason why these are not also affected by the dynamics in MDM2 structure. One interesting aspect is to exploit the allosteric nature of MDM2 for novel approaches for therapeutic drug development aimed at controlling p53 activity. Traditionally, this has been restricted to interfere with the p53-MDM2 interface, and drugs such as nutlins have been developed and tested in clinical trials aimed at activating wild-type p53 by releasing MDM2-mediated suppression. However, targeting certain functions of MDM2 toward p53 and, for that matter, toward other interacting partners can alternatively be developed by taking advantage of the dynamics of MDM2 (Fig. 3). For example, mimicking a conformation of MDM2 that resembles its RNA-bound stage could potentially render MDM2 a positive regulator of p53 and not just prevent its degradation of p53. Similarly, drugs that fit into its ATP pocket within the RING domain might alter the specific activity of MDM2 as a protein foldase. It has been shown that zinc-bound forms of MDM2 are activated for binding to the oncoprotein HSP90,46 and this HSP90-MDM2 “oncocomplex” might be linked in the myc-dependent tumorgenesis as-sociated with the MDM2 allele with mutation in the zinc finger.47 Identifying inhibitors or mimetics of such protein-protein interactions between HSP90: MDM2, MDM2:RNA, and other interactions requires techniques that are under development and that will allow desired structural alterations to be monitored on a quantitative high-throughput basis. A current restriction in targeting protein-protein interactions has been that “druggable” targets are validated based on the interface of the 2 proteins in question and whether this is deemed sufficient to find small molecules that can compete. This has been a major concern and a frequent argument to prevent new programs from being launched that aim at modifying protein-protein interactions. But when targeting allosteric proteins like MDM2, other possibilities open up, and it might be worthwhile to keep an open mind and not restrict oneself to the PPI interface. However, it is difficult to predict how small molecules will give a desired effect via modifications of protein dynamics, and for this reason, new ideas and techniques need to be developed. For example, nanoparticle assays that measure oligomerization of MDM2 can identify novel types of ligands that are not possible using assays that do not sense the oligomerization state of MDM2.48 Thus, in-depth biochemical and cellular analysis of the dynamics of 2 proteins interacting will not only identify mechanisms of how to identify keys to unlock interactions but also modify one function while retaining others. Such assays can help to identify compounds that can “fine-tune” the p53-MDM2 axis and that can selectively and independently control the 3 key biological outcomes linked to MDM2-mediated control of p53 activity: aging, cancer suppression, and longevity. One can also argue that allosteric protein-protein interactions are best studied in their native environment in the cell. For this purpose, the Bioluminescence Resonance Energy Transfer (BRET) assay is an alternative.49 This assay is not at the level where one can determine changes in certain domains but allows detection of the interaction between 2 proteins in live cells and has been used to look at the dynamics of the p53-MDM2 interphase using nutlins.50 This assay also has been successfully applied in yeast, which might allow larger screening assays using compounds or peptide aptamers.51 Importantly, as the p53-MDM2 axis forms an attractive target for cancer therapies, this field will continue to attract industrial and academic scientists to develop new techniques and concepts that will not only serve to modify the p53 pathway but will spill over to benefit other fields as well.
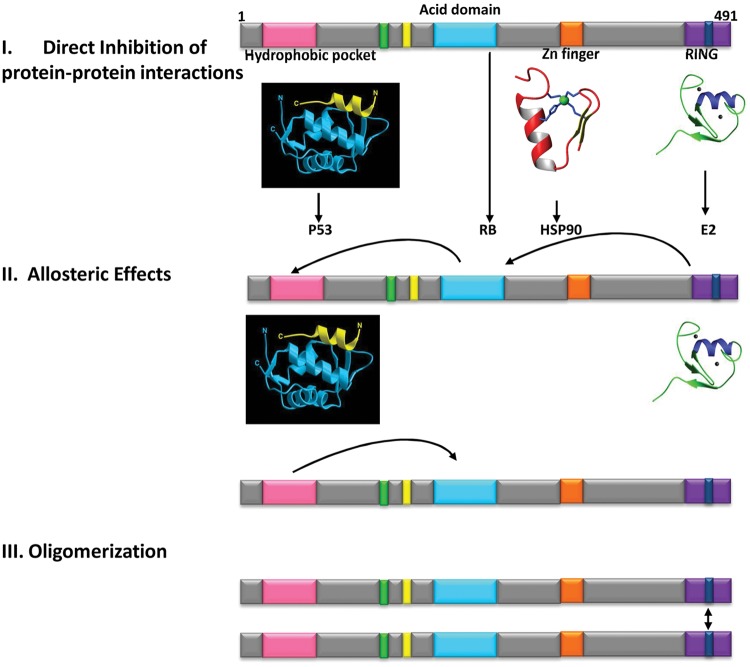
Figure 3.
Understanding protein-protein contacts in the MDM2 protein to develop novel small-molecule regulatory screens to identify molecules that modify one or several MDM2 functions. Three distinct concepts surrounding how to target MDM2 have been developed from biochemical, biophysical, and cellular analyses. I. Direct inhibition of protein-protein interactions. MDM2 is composed of at least 4 distinct functional domains that regulate its activity: the N-terminal regulatory domain, the central acidic domain, a zinc finger domain, and the C-terminal RING domain. Each domain is implicated in various functions of MDM2. This has either provided novel small-molecule regulators (like Nutlin-3 targeting the N-terminal domain52) or offered potential for novel small-molecule developments highlighted by the identification of specific peptide-mimetic inhibitors of the acidic domain,17,53 stimulation of zinc-dependent allosteric peptide interactions that might alter interaction with pro-oncoproteins such as HSP90,46,47 and small molecules or peptide-mimetics that target the E3-E2 interfaces.54 II. Allosteric changes in MDM2. Mutation of the RING domain can alter the affinity of the N-terminal domain and/or the acidic domain for its ligand,55 and conversely, mutating the N-terminal lid motif can stimulate acidic domain interactions.56 In addition, biophysical studies demonstrated direct allosteric effects on intrinsic fluorescence of amino acids within the central acidic domain upon perturbation of either the RING domain or the N-terminal MDM2-lid. Together, these studies highlight the concept that novel information can be acquired on ligand binding functions using full-length MDM2 and by taking into consideration such allosteric properties. Accordingly, novel protein-protein interaction assays can be envisioned. III. Dynamic protein-protein interactions. A novel molecular chaperone function of MDM2 involving refolding of denatured luciferase, p53 conformational changes, or perturbing E2F1 conformational perturbation provides a complex multidomain non-ubiquitination function for MDM257,58 that might provide novel drug leads that modify MDM2 foldase function. Similarly, the stimulation of MDM2 by RNA on protein-protein interactions,46 p53 ubiquitination,59 or p53 protein synthesis60 provides another distinct complex assay that might provide druggable opportunities. A nanoparticle assay was designed that measures “oligomerization” of a protein toward its ligand, and using this assay, novel RING-dimerization domain inhibitory peptides have been identified supporting the concept that novel assays incorporating allostery can produce new ideas on developing drug leads that target MDM2.48
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101), La Ligue Contre le Cancer and CRUK.
References
- 1. Tyner SD, Venkatachalam S, Choi J, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53 [DOI] [PubMed] [Google Scholar]
- 2. Gannon HS, Donehower LA, Lyle S, Jones SN. Mdm2-p53 signaling regulates epidermal stem cell senescence and premature aging phenotypes in mouse skin. Dev Biol. 2011;353:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maier B, Gluba W, Bernier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17:201–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hara MR, Kovacs JJ, Whalen EJ, et al. A stress response pathway regulates DNA damage through beta2-adrenoreceptors and beta-arrestin-1. Nature. 2011;477:349–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, et al. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matheu A, Maraver A, Klatt P, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–9 [DOI] [PubMed] [Google Scholar]
- 8. Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6 [DOI] [PubMed] [Google Scholar]
- 9. Kastan MB, Zhan Q, el-Deiry WS, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–97 [DOI] [PubMed] [Google Scholar]
- 10. Lee IH, Kawai Y, Fergusson MM, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Itahana K, Mao H, Jin A, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–66 [DOI] [PubMed] [Google Scholar]
- 12. Zhu J, Zhou W, Jiang J, Chen X. Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J Biol Chem. 1998; 273:13030–6 [DOI] [PubMed] [Google Scholar]
- 13. Yin Y, Stephen CW, Luciani MG, Fahraeus R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol. 2002;4:462–7 [DOI] [PubMed] [Google Scholar]
- 14. Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000;20:8458–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shloush J, Vlassov JE, Engson I, et al. Structural and functional comparison of the RING domains of two p53 E3 ligases, Mdm2 and Pirh2. J Biol Chem. 2011;286:4796–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–73 [DOI] [PubMed] [Google Scholar]
- 17. Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell. 2006;23:251–63 [DOI] [PubMed] [Google Scholar]
- 18. Iyappan S, Wollscheid HP, Rojas-Fernandez A, et al. Turning the RING domain protein MdmX into an active ubiquitin-protein ligase. J Biol Chem. 2010;285:33065–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–8 [DOI] [PubMed] [Google Scholar]
- 20. Tang X, Orlicky S, Lin Z, et al. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–76 [DOI] [PubMed] [Google Scholar]
- 21. Candeias MM, Malbert-Colas L, Powell DJ, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10:1098–105 [DOI] [PubMed] [Google Scholar]
- 22. Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–11 [PubMed] [Google Scholar]
- 23. Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J. 1995;14:4442–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu L, Benchimol S. Participation of the human p53 3′UTR in translational repression and activation following gamma-irradiation. EMBO J. 1997;16:4117–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63 [DOI] [PubMed] [Google Scholar]
- 26. Mazan-Mamczarz K, Galban S, Lopez de, Silanes I, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A. 2003;100:8354–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang DQ, Halaby MJ, Zhang Y. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene. 2006;25:4613–9 [DOI] [PubMed] [Google Scholar]
- 28. Gajjar M, Candeias MM, Malbert-Colas L, et al. The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell. 2012;21:25–35 [DOI] [PubMed] [Google Scholar]
- 29. Maya R, Balass M, Kim ST, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ray PS, Grover R, Das S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006;7:404–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Candeias MM, Powell DJ, Roubalova E, et al. Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene. 2006;25:6936–47 [DOI] [PubMed] [Google Scholar]
- 32. Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–37 [DOI] [PubMed] [Google Scholar]
- 33. Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 2009;15:363–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mayer C, Grummt I. Cellular stress and nucleolar function. Cell Cycle. 2005;4:1036–8 [DOI] [PubMed] [Google Scholar]
- 35. Scheer U, Hock R. Structure and function of the nucleolus. Curr Opin Cell Biol. 1999;11:385–90 [DOI] [PubMed] [Google Scholar]
- 36. Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poyurovsky MV, Jacq X, Ma C, et al. Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Mol Cell. 2003;12:875–87 [DOI] [PubMed] [Google Scholar]
- 38. Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Niedick I, Froese N, Oumard A, et al. Nucleolar localization and mobility analysis of the NF-kappaB repressing factor NRF. J Cell Sci. 2004;117:3447–58 [DOI] [PubMed] [Google Scholar]
- 40. Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci U S A. 1999;96: 6937–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xirodimas DP, Chisholm J, Desterro JM, Lane DP, Hay RT. P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2. FEBS Lett. 2002;528:207–11 [DOI] [PubMed] [Google Scholar]
- 42. Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: a multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair (Amst). 2009;8:1215–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine AJ. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol Med. 1996;2:439–51 [PMC free article] [PubMed] [Google Scholar]
- 45. Maslon MM, Hupp TR. Drug discovery and mutant p53. Trends Cell Biol. 2010;20:542–55 [DOI] [PubMed] [Google Scholar]
- 46. Burch L, Shimizu H, Smith A, Patterson C, Hupp TR. Expansion of protein interaction maps by phage peptide display using MDM2 as a prototypical conformationally flexible target protein. J Mol Biol. 2004;337:129–45 [DOI] [PubMed] [Google Scholar]
- 47. Macias E, Jin A, Deisenroth C, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell. 2010;18:231–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Robson AF, Hupp TR, Lickiss F, Ball KL, Faulds K, Graham D. Nanosensing protein allostery using a bivalent mouse double minute two (MDM2) assay [published online May 3, 2012]. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1116637109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Milligan G. Applications of bioluminescence- and fluorescence resonance energy transfer to drug discovery at G protein-coupled receptors. Eur J Pharm Sci. 2004;21:397–405 [DOI] [PubMed] [Google Scholar]
- 50. Mazars A, Fahraeus R. Using BRET to study chemical compound-induced disruptions of the p53-HDM2 interactions in live cells. Biotechnol J. 2010;5:377–84 [DOI] [PubMed] [Google Scholar]
- 51. Corbel C, Wang Q, Bousserouel H, et al. First BRET-based screening assay performed in budding yeast leads to the discovery of CDK5/p25 interaction inhibitors. Biotechnol J. 2011;6:860–70 [DOI] [PubMed] [Google Scholar]
- 52. Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8 [DOI] [PubMed] [Google Scholar]
- 53. Midgley CA, Desterro JM, Saville MK, et al. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–23 [DOI] [PubMed] [Google Scholar]
- 54. Kar G, Keskin O, Nussinov R, Gursoy A. Human proteome-scale structural modeling of E2-E3 interactions exploiting interface motifs. J Proteome Res. 2012;11:1196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wawrzynow B, Pettersson S, Zylicz A, et al. A function for the RING finger domain in the allosteric control of MDM2 conformation and activity. J Biol Chem. 2009;284:11517–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Worrall EG, Wawrzynow B, Worrall L, Walkinshaw M, Ball KL, Hupp TR. Regulation of the E3 ubiquitin ligase activity of MDM2 by an N-terminal pseudo-substrate motif. J Chem Biol. 2009;2:113–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stevens C, Pettersson S, Wawrzynow B, et al. ATP stimulates MDM2-mediated inhibition of the DNA-binding function of E2F1. FEBS J. 2008;275:4875–86 [DOI] [PubMed] [Google Scholar]
- 58. Wawrzynow B, Zylicz A, Wallace M, Hupp T, Zylicz M. MDM2 chaperones the p53 tumor suppressor. J Biol Chem. 2007;282:32603–12 [DOI] [PubMed] [Google Scholar]
- 59. Shimizu H, Burch LR, Smith AJ, et al. The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J Biol Chem. 2002;277:28446–58 [DOI] [PubMed] [Google Scholar]
- 60. Candeias MM, Malbert-Colas L, Powell DJ, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10:1098–105 [DOI] [PubMed] [Google Scholar]