Abstract
Cancer cells often have high expression of Mdm2. However, in many cancers mdm2 is alternatively spliced, with more than 40 mRNA variants identified. Many of the alternative spliced mdm2 mRNAs have the potential to encode truncated Mdm2 isoforms. These putative Mdm2 isoforms can theoretically increase the diversity of the cancer proteome. The 3 best characterized are Mdm2-A, Mdm2-B, and Mdm2-C. As described in this review, the exogenous expression of these isoforms results in paradoxical phenotypes of transformation-associated growth as well as the inhibition of growth. Interestingly, these Mdm2 isoforms contribute tumor-promoting capacity in p53-null backgrounds. Herein we describe how alternative splicing of mdm2 may result in Mdm2 protein products that alter signal transduction to promote tumorigenesis. The tumor promoting capacity of Mdm2 isoforms is discussed in the context of functions that do not require the inhibition of p53. When N-terminal portions of Mdm2 are missing, the biochemical functions encoded by exon 12 are proposed to become more important. This may result in growth promoting functions when wild-type p53 is absent or compromised. The p53-independent tumor promoting activity of Mdm2 is proposed to result from C-terminal biochemical contributions of DNA binding, RNA binding, nucleolar localization, and nucleotide binding.
Keywords: Mdm2, splicing, proteome, cancer
Introduction
In cancer cells, the Mdm2 protein often contributes to altered molecular programs that increase growth-promoting signals and decrease cell death signals. An important growth inhibitory program in normal cells is the p53 pathway. A central negative regulator of p53 is the Mdm2 protein, so it is not surprising that Mdm2 was first identified by its physical interaction with p53.1,2 Mdm2 is well known for its canonical function; the protein inhibits p53 through a negative feedback loop in which p53 activates the expression of mdm2 and Mdm2 acts as an E3 ubiquitin ligase to promote the degradation of p53.3
High expression of Mdm2 is seen in diverse types of human cancer; however, the predictive value of Mdm2 as an oncology biomarker is controversial.4 The complexity of Mdm2 as a biomarker is partly due to the existence of numerous alternatively spliced mdm2 transcripts.5-10 Alternative splicing of mdm2 mRNAs in cancers has the potential to vastly increase the cancer-associated proteome.11,12 Cancer specific splicing events are seen for a wide range of tumor associated proteins.11 This review explores the putative polypeptide isoforms of Mdm2 that are contributed to the cancer proteome and discusses how Mdm2 isoform diversity might support oncogenic transformation.
Alternative Splicing of Mdm2: Increasing Mdm2 Proteome Diversity
Human cancers often have high levels of alternatively spliced mdm2 mRNAs.5-10 The wide variety of alternative spliced mdm2 mRNAs are well described in a number of excellent reviews.10,13 Whereas the mRNAs of more than 40 mdm2 variants have been documented, significantly less has been discussed about the putative function of the endogenous truncated polypeptide isoforms. The full-length Mdm2 protein is a 491 amino acid polypeptide; however, it is doubtful that the overexpression of Mdm2 in human cancers consists solely of the full-length Mdm2 isoform (Mdm2-FL).
In 1996, the Lunec laboratory identified multiple-sized mdm2 transcripts from a group of snap-frozen primary tumor samples and named them mdm2-a, mdm2-b, mdm2-c, mdm2-d, and mdm2-e. 5 The mdm2-a, mdm2-b, and mdm2-c mRNAs result from bona fide alternative splicing. Their translation products will be focused on in this review to develop the concept that Mdm2 isoforms increase cancer proteome diversity. The detection of endogenous polypeptides resulting from alternatively spliced mdm2 has not been published. We delivered an oral report at the 2011 Mdm2 meeting documenting the detection of endogenous Mdm2-C in human patient derived liposarcomas and the human breast cancer cell line T47D (Okoro and Bargonetti, manuscript in preparation). We detected endogenous Mdm2-C by using a polyclonal antibody produced to the polypeptide sequence of the splice junction of exons 4 and 10. Our detection of Mdm2-C can be used as evidence to promote the hypothesis that Mdm2 isoforms contribute to cancer proteome diversity.
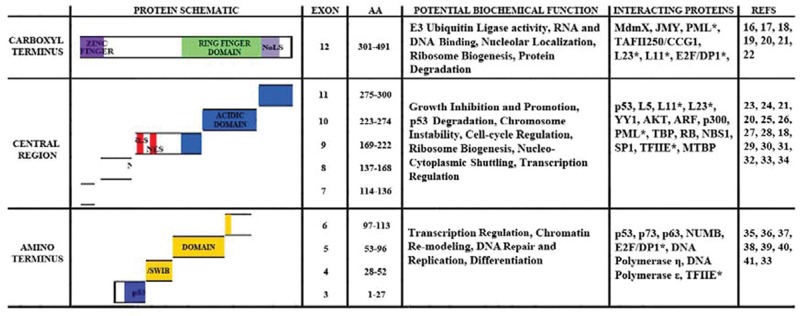
What suggests that alternatively spliced messages of mdm2 encode proteins that control growth regulatory switches? Two formidable examples of alternative splicing support a hypothesis that cancer-related alternative splicing of mdm2 can be a profound regulator. They are the sex-determination regulatory switch in Drosophila (involving sxl, tra, msl2, dsx, and fru) and the activated hair cell tuning switch in humans (controlled by the calcium-activated potassium channel gene, slo).12 Additionally, a recently discovered splice variant of traditional FOXP1 that controls human embryonic stem cell pluripotency and reprogramming suggests that mRNA splicing is an important component of growth control.14,15 The growth regulating embryonic stem cell specific FOXP1 (FOXP1-ES) places alternative splicing at the top of the hierarchy for controlling pluripotency and reprograming.14 The structure of the mdm2 gene, its multiple alternatively spliced products, the domains present in the amino acid sequence, and the numerous Mdm2-interacting proteins suggest important p53-independent and p53-dependent growth regulatory properties of the Mdm2 isoforms (Fig. 1).16-41
Figure 1.
Domain analysis of Mdm2 coding exons including potential biochemical functions. The Mdm2 protein is shown as 3 regions. The carboxyl, central, and amino terminal regions are shown with their domains. From C-terminus to N-terminus, the domains represented are the Nucleolar localization signal (NoLS), the RING finger, the Zinc finger, the acid domain, the nuclear localization (NLS) and export signals (NES), and the SWI/SNF Chromatin Remodeling (SWIB)/p53 binding domain. The amino acid (AA) sequence numbers are shown for the exons. The potential biochemical functions of each region are given. Finally, the interacting proteins are listed and references for these are provided. *Proteins that bind in overlapping regions. National Center for Biotechnology Information (NCBI) blast results were used as a reference for Mdm2 binding proteins.
The functions of putative Mdm2 isoforms require increased study. The mdm2 gene contains 12 exons. Depending on which promoter is used for the gene’s transcription, the mature message contains either exon 1 or exon 2.8 Exons 1 and 2 produce noncoding mRNA sequences. The coding sequences are found in exons 3 through 12 and encode the full length Mdm2 protein (Mdm2-FL) (see Fig. 1). Most of the mdm2 splice variants found in cancer have exon 12, which contains one-third of the protein coding sequence. This exon encodes the Zinc finger region, the nucleolar localization signal (NoLS), and the RING finger domain. In alternatively spliced isoforms, however, key information regions are lost. An important region removed from the central domain is the acidic portion of the coding sequence, which is vital for interacting with p53 and is required for p53 proteolysis.23,42,43 A region removed from the amino terminus of Mdm2 has homologous sequence to the SWI/SNF chromatin remodeling domains in addition to a p53 interacting domain.44 The smallest spliced variant found in human cancers is mdm2-b, and it contains a combination of exons as follows: Promoter 1 (P1) firing results in a transcript that contains exon 1 + exon 3 + exon 12 = mdm2-b transcript; promoter 2 (P2) gives a transcript that contains exon 2 + exon 3 + exon 12 = mdm2-b. The Mdm2-B polypeptide comes from translation of exons 3 and 12. This Mdm2-B isoform is able to cause tumors in mice,45 and mRNA for mdm2-b is found in many different types of cancer.5,6 The Mdm2-A isoform would arise from translation of exon 3 + exon 10 + exon 11 + exon 12 and is able to cause tumors in mice.46 The Mdm2-C isoform would arise from the translation of exon 3 + exon 4 + exon 10 + exon 11 + exon 12 and has not yet been examined in transgenic mice.
The association of correlated cancer phenotypes extends the description of alternative splicing of mdm2 to tumor tissues. High levels of alternative mdm2 splicing are seen in tissue samples from ovarian and bladder carcinomas, some types of leukemia, breast carcinoma, soft tissue sarcoma, Hodgkin’s lymphoma, glioblastoma, rhabdomyosarcomas, liposarcomas, and lung carcinomas.5,9,10,13,47-51 Mouse model information suggests that the tumor promoting capacity of Mdm2 does not require inhibition of wild-type p53. Exogenous expression of Mdm2-FL in mice was documented in 1998 to predispose the animals to sarcomas in a p53-independent manner.52 Exogenous expression of some, but not all, murine mdm2 splice variant isoforms expressed in transgenic mouse models also predisposes p53 null mice to tumor formation.53 An additional set of p53 null mice data showed that exogenous expression of Mdm2-A promotes tumor formation.46 The fact that Mdm2-FL protein and truncated isoforms of Mdm2 promote tumorigenesis in the absence of p53 indicates an intricate Mdm2 signal transduction profile. The 12th exon of Mdm2 might be responsible for the p53-independent tumor promoting outcomes. It encodes at least one-third of the amino acids in all Mdm2 isoforms and has the potential to participate in RNA binding, DNA binding, E3 ubiquitin ligase activity, nucleolar localization, and chaperone activity (see Fig. 1).54-60 Therefore, it stands to reason that exon 12 encodes some of the non-p53 targeting tumor promoting biochemical functions.
The possibility for Mdm2 isoform diversity exists because of the large array of alternatively spliced mdm2 transcripts under stressful and/or pathological conditions.13 DNA lesions that correlate with helix distortions promote alternative splicing of mdm2, and their production is not a passive outcome but rather an active decision.61,62 Stress induces the activation of the mdm2 P2 promoter by p53, Sp1, and Estrogen Receptor (ER).63-66 The mdm2 mRNAs transcribed from the downstream (P2) promoter are translated into proteins more efficiently than those from the upstream (P1) promoter.67 However, transcripts resulting from both the P1 and P2 promoters yield UV-mediated alternative splicing of the downstream exons.61 Although the health of the cell has more influence on alternative splicing outcomes than promoter usage, a transcript from the P2 promoter should be translated more effectively.
Stress-induced mdm2 mRNAs existing in aggressive tumors suggest that the genomic instability in tumors drives mdm2 alternative splicing. The biological function of these mdm2 mRNAs is most likely to encode the proteomic diversity that is required when a cell encounters a stressful situation. Mdm2-A and Mdm2-B isoforms, resulting from stress-induced mdm2 mRNAs, inhibit Mdm2-FL activity and activate the p53-dependent cell death pathway.46,62,68,69 When wild-type p53 function is compromised (by mutation or the production of oncogenic products), the mdm2 splice variants may take on new transforming roles. The correlation of increased alternative spliced mdm2 mRNAs with aggressive cancers supports the need for the putative Mdm2 truncated isoforms to maintain cell survival following a genomic stressful situation. The spliced variants of mdm2 may be co-opted for growth promoting functions when wild-type p53 function is compromised.
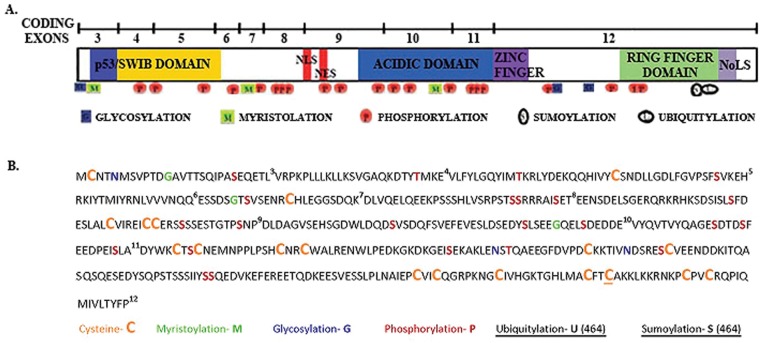
The detection of endogenous Mdm2 truncated isoforms has been a challenge due to the lack of specific antibodies to detect such isoforms.13 Antibodies to Mdm2 (see Table 1) are able to detect multiple isoforms of Mdm2, but exactly how these relate to the splice variant isoforms is not clear.35,64,70 Additionally, the Mdm2 protein migrates larger than the expected size in SDS-PAGE. The reasons for this discrepancy may be due to potential post-translational modifications and/or acidic region contributions (see Fig. 2).35 For example, Mdm2-FL has a predicted molecular weight of 56 kDa but has an apparent molecular weight of 90 kDa.35 The splice variants show similar aberrant mobilities. Mdm2-A has a predicted molecular weight of 30 kDa but has an apparent molecular weight of 75 kDa.58 This retarded gel mobility of Mdm2, and the challenge to determine what forms of Mdm2 the antibodies detect, have limited the identification of proposed Mdm2 isoforms. Mdm2 is cysteine-rich, with 18 Cys in the sequence (Fig. 2). A total of 13 Cys are encoded by exon 12. The Mdm2-FL protein can potentially form 9 disulfide linkages. We see considerable gel migration discrepancy for Mdm2-FL and Mdm2-C that is changed after treatment of our samples with strong reducing agents (Okoro and Bargonetti, manuscript in preparation). The changes in the Mdm2 disulfide scaffold in the isoforms most likely contribute to the proteome diversity. Detection of Mdm2 and its isoforms by immunohistochemistry (IHC) and immunofluorescence (IF) does not encounter mobility problems. We have observed endogenous Mdm2-C in the cytoplasm and nucleolus of cancer cell lines and liposarcoma tissue using our newly developed specific polyclonal antibody (Okoro and Bargonetti, manuscript in preparation). This gives us cause to speculate that endogenous Mdm2 isoforms confer diversity to the proteome in cancer tissue.12,14,58
Table 1.
Commonly Used Mdm2 Antibodies
| Epitope region | Exons included | Antibodies | Some commercial sources |
|---|---|---|---|
| 383-491 | 12 | 4B11a | EMD Millipore |
| 5B10a | |||
| 294-339 | 11-12 | 2A10a | Abcam |
| EMD Millipore | |||
| Thermo-Scientific Pierce Antibodies | |||
| 100-320 | 6-12 | D-7 | Santa Cruz Biotechnology, Inc. |
| 153-222 | 8-9 | 2A9a | EMD Millipore |
| 3G9a | |||
| 154-167 | 8 | SMP14 | Abcam |
| EMD Millipore | |||
| Sigma-Aldrich | |||
| Santa Cruz Biotechnology, Inc. | |||
| Thermo-Scientific Pierce Antibodies | |||
| 26-169 | 3-8 | IF2 | EMD Millipore |
| Invitrogen | |||
| 19-50 | 4 | 4B2a | EMD Millipore |
| 1-50b | 3-4 | N-20 (polyclonal) | Santa Cruz Biotechnology, Inc. |
Note: Antibodies are monoclonal unless otherwise stated.
Clones originated from the laboratory of Arnold Levine35 that are commercially available.
An approximate epitope region given by company.
Figure 2.
Putative post-translational modifications on the Mdm2 polypeptide. (A) The 3 major regions of Mdm2. (B) The protein coding sequence of Mdm2 with the putatively modified amino acids. All Cysteine (C) residues are colored orange and represent potential residues for the formation of disulfide bonds. Some residues are colored to indicate sites of potential modifications: Asparagine (N)–Glycosylation; Glycine (G)–Myristoylation; Serine (S) and Threonine (T)–Phosphorylation; and Cysteine at position 464 (C)–ubiquitylation/sumoylation. ExPASY.org/prosite program was used to determine the potential modified residues.
The putative Mdm2 isoforms have lost a significant portion of the N-terminal p53 binding domain and result predominantly from exon 12 (Fig. 1). This suggests an increased role of RNA regulation for truncated Mdm2 isoforms because they should maintain RNA-binding activity.55,57,58 Exogenously expressed Mdm2-A and Mdm2-B can localize to the nucleoplasm, although they no longer contain the nuclear localization signal.71 They have also been detected in the cytoplasm where binding to RNA could impart a major regulatory property.9,62,72 Due to the fact that no tumor tissue has been compared for endogenous Mdm2 isoform polypeptides and all tumor studies to date have examined mRNA in cancer, it is difficult to assess the prognostic implications for the protein isoforms. Moreover, many of the Mdm2 antibodies used for tumor analysis do not interact with the Mdm2 isoforms that would result from the spliced variants (Table 1). Therefore, the published Mdm2 immunohistochemistry data of cancers give a low representation of the cancer specific proteomic diversity. The predictive capability of Mdm2 isoforms markers is missed when we look at total “Mdm2 positivity.” The antibody reagents in the field need to be improved so we can move forward with better prognostic indicators.
Mdm2 Isoforms Deliver Multiple Regulatory Functions
When Mdm2 is overexpressed in cancer, there is a disruption in wild-type p53 signal transduction that leads to increased cell growth.73 However, there is strong evidence that not all Mdm2 oncogenic activity comes from targeting p53 for degradation and/or inhibiting the p53 transcriptional activity.74,75 Many chemotherapeutics directed at Mdm2 target the p53-Mdm2 interaction (e.g., the Nutlins) and are directed to the N-terminal p53 binding pocket of Mdm2.76 This region is encoded by exons 3, 4, and 5 of the mdm2 transcript and is truncated in most mdm2 splice variants. Other drugs under development target the E3-ubiquitin ligase activity of Mdm2 via the RING finger domain.73 However, in the absence of the central acidic domain, the E3 ligase activity of Mdm2 toward p53 is lost.42 Therefore. as we consider the biochemical activity of the splice variants and their missing regions, we should contemplate what activity would be blocked by the new era of Mdm2 targeted chemotherapeutics.
The Mdm2 protein has several conserved domains with characterized biochemical activities.77 We will consider the activities resulting from the carboxyl terminus first because the spliced variants all contain exon 12. The carboxyl terminus of Mdm2 has many potential biochemical activities (Fig. 1). The Zinc finger domain is encoded by exon 12 and possesses a structure in solution that demonstrates potential DNA and RNA binding activity.59 No documented biochemical activity data exist for the Mdm2 Zinc finger domain; however, the solution structure is similar to that seen for ribosomal proteins, transcription factors, DNA replication factors, protein-DNA interactors, and protein-RNA interactors.59 There are documented biochemical activity data for the C-terminal RING finger domain of Mdm2 (also encoded by exon 12). The RING finger domain is important for Mdm2 binding to DNA and RNA.55,57,58 In addition, the RING finger domain contains a nucleolar localization signal.71 This domain also promotes degradation of proteins including p53, Mdm2, and MdmX.3,78 However, the C-terminus alone is not sufficient to promote p53 proteolysis. The interaction of Mdm2’s N-terminus and central acidic domain is also required for p53 degradation42 (Fig. 1). Mdm2 growth inhibitory domains are encoded within exons 9 and 10 and part of 12.79 The nuclear localization and export signals, encoded by exon 9, are responsible for Mdm2 nucleo-cytoplasmic shuttling.3 Some examples of proteins that interact with the central region, in addition to p53, are p300 and ribosomal protein L5.3 At the N-terminus is the p53 binding domain, encoded by exons 3, 4, and 5.35 The Mdm2 amino acid sequence 10 to 139 can bind to both the N- and C-termini of p53, contributing significantly to p53 regulation.80
The multiple biochemical activities of the Mdm2 C-terminus, in the absence of significant central regions deleted from the splice variants, could be responsible for the transforming activity of truncated Mdm2 isoforms. Mouse models support the theory of transforming activity of truncated Mdm2, as a series of mouse mdm2 variants lacking the central region placed into Eµ-myc transgenic mice promote tumorigenesis.53 The expression of these mdm2 splice variants in mice accelerates lymphomagenesis and promotes aggressive lymphomas.53 When p53 heterozygous mice are crossed with Mdm2-A transgenic mice, the resultant mice develop aggressive mammary tumors.46 Furthermore, in a p53 null background, addition of Mdm2-A enhances transformation of mouse embryonic fibroblasts (MEFs) and alters the tumor spectrum in p53-null and Arf-null mice.46 In p53-, ARF-, and Rb-null MEFs, Mdm2-B increases cell proliferation.45 Mdm2-B confers anti-apoptotic properties due to an increase in protein levels of the p65 RelA subunit of NFκB. Tissue restricted expression of the Mdm2-B isoform in mice induces spontaneous tumor formation that correlates with increased p65 RelA subunit expression in the tumors.45 Although the mouse models do not recapitulate all the data seen in human cell culture, they firmly support the finding that Mdm2 isoforms have p53-independent transforming activity. Thus, the functions of these Mdm2 isoforms may be tissue and/or cancer-type specific, and, depending upon the genetic background of the cancer, the Mdm2 isoforms may behave differently.
The large array of protein-protein binding partners for Mdm2 will vary in their activity as the concentration of spliced variant isoforms is increased (Fig. 1). Functions of these binding partners place Mdm2 in pathways involved in cell cycle progression, differentiation, DNA synthesis, ribosome biogenesis, transcription, and membrane trafficking.74 The putative Mdm2 protein isoforms lose key binding partners. The removal of Mdm2 growth inhibitory domains may also contribute to Mdm2 isoform associated proliferation seen in cancers.79 The retention of the C-terminus including the RING finger domain may be the most significant indicator that the p53-independent activities of the Mdm2 isoforms are what contribute to the functional diversity in cancers.58
Mdm2 Isoforms and Molecular Partners in Cancer
A way to consider the functions of putative Mdm2 isoforms in cancer is to analyze what regions they maintain and what regions are missing. The majority of the known mdm2 splice variants that would encode protein are devoid of the NLS and NES domains but retain the nucleolar localization sequence (NoLS).13 The cellular localization of Mdm2 is dependent on post-translational modifications at residues in the acidic domain as well as at the NLS and NES regions.81 We have examined the potential post-translational modification of Mdm2 using the computer program ExPASY.org prosite, and we find that there are more modifications than have been described in the literature (Fig. 2). The combinatorial changes in these modifications coupled with the deletion of central Mdm2 portions of the protein could substantially influence proteome diversity and biochemical functions.
The Mdm2 protein interacts with p53 using 3 regions: the amino terminus binding pocket, the central acidic domain, and a region in the carboxyl terminus.80 Mdm2 isoforms that lack sequence encoded by exons 3-5 will be unable to promote p53 degradation, but this would not necessarily inhibit p53 protein binding. Cancers and cell lines with high levels of alternatively spliced mdm2 mRNA often have stabilized p53 protein.47,82 The reason for p53 stability in these cancers could be the due to the “nanny” functions of Mdm2 isoforms. Nanny proteins help protect intrinsically disordered proteins, known as IDPs, from degradation.83 Full-length Mdm2 can protect p53 in the absence of its ability to target it for degradation because p53 is an IDP.84 We have observed that the Mdm2 isoform Mdm2-C, when exogenously co-expressed with p53, is unable to degrade p53 compared with the Mdm2-FL (Okoro and Bargonetti, manuscript in preparation). Based on this observation, we hypothesize that the N-terminally truncated isoforms might also stabilize oncogenic mutant p53.
Mdm2 also functions as a chaperone, singly or in combination with HSP90, and assists in p53 folding and subsequent DNA binding.56 Mdm2 promotes a change in p53 conformation using the RING finger domain to result in cytoplasmic shuttling.85 Truncated Mdm2 isoforms retaining the RING finger domain should have the ability to act as molecular chaperones. However, it is unknown whether the amino terminus of Mdm2 is required for the chaperone activity of Mdm2. The E3 ligase activity of Mdm2 is required for chaperone activity to ubiquitinate p53 at the central domain.43 Whether Mdm2 isoforms retain E3 ligase activity is yet to be determined.
The interacting regions for many Mdm2 protein-binding partners that participate in p53-independent functions are lost in the truncated isoforms.31,74 However, how loss of these protein interactions will influence growth outcomes is not clear. Some examples are shown in Figure 1. E2F1 is such an example as it interacts with Mdm2 through the amino terminus and the RING finger domain.22,39 Because the RING domain of Mdm2 also binds to E2F, it is difficult to determine what the final outcome of truncated isoforms would do to the Mdm2-E2F interaction. Mdm2 also binds to the DNA repair protein NBS1 using a region absent in truncated isoforms.86 The interaction with NBS1 induces chromosome instability75 and gives B-cells a growth advantage.87 The region of Mdm2 that binds to NBS1 is largely missing in Mdm2 isoforms, implying that overexpression of Mdm2 isoforms will not result in genomic instability. More research will be needed to completely understand the different molecular mechanisms that deliver variable outcomes.
In its carboxyl terminal region, Mdm2 possesses a Zinc finger domain that has a consensus sequence similar to that found in proteins associated with transcription and translation regulation.59 Although many Mdm2 isoforms retain this domain, its function is still unknown. The RING finger domain of Mdm2 binds to DNA and contains a Walker A motif that is common to ATP/GTP binding proteins.88 Mdm2-FL shows some sequence specificity for the choice of binding to DNA and RNA molecules.57 The RING finger domain of Mdm2 also activates histone ubiquitylation, leading to transcriptional repression.89 It has not been determined whether Mdm2 truncated isoforms have DNA and RNA binding specificity. The RING finger is maintained in the Mdm2 truncated isoforms and therefore most likely influences DNA- and RNA-mediated events. Interestingly, cells that overexpress Mdm2 via a single nucleotide polymorphism (mdm2 SNP 309) in the second promoter region of the mdm2 gene generate a spectrum of mdm2 splice variants that are unique from non-Mdm2 overexpressing cells.90 Homozygous SNP 309 cancer cells also have repressed activation of p53 target genes in the presence of wild-type p53 protein.91 Homozygous SNP 309 cells maintain Mdm2 on the chromatin of p53-target genes even after DNA damage. This suggests that Mdm2 isoforms may be present with p53 on the chromatin and thereby block the activation of transcription needed for tumor suppression.91
Mdm2 binds to RNA using the RING finger domain.55 When Mdm2 binds to p53 mRNA, it promotes the translation of p53 protein60,92 and impairs the E3 ligase activity.93 Interestingly, the region of p53 mRNA that binds to the Mdm2 RING domain is the same p53 sequence that encodes the N-terminal amino acids that interact with the Mdm2 protein.60,94 An Mdm2 isoform, p76, lacking the p53-binding domain, binds to p53 mRNA and promotes efficient translation of the mRNA but does not promote p53 protein degradation.60 This implies that N-terminal truncated Mdm2 isoforms might be better translation effectors than their Mdm2-FL counterpart.
The positive role of Mdm2 on p53 mRNA translation is activated by the Ataxia Telangiectasia Mutated (ATM) kinase.92 ATM phosphorylates Mdm2 at Ser 395, which increases Mdm2-p53 mRNA binding activity.92 In theory, Mdm2 truncated isoforms should be capable of RNA binding that is activated by ATM. However, Mdm2 isoforms have not been experimentally examined for their influence on mRNA translation. The phosphorylation of Mdm2 by ATM kinase facilitates the transport of Mdm2 to the nucleolus, where it binds to the p53 mRNA.92 Theoretically, Mdm2 can influence the cancer proteome by interacting with many different mRNA molecules to activate, or inhibit, the translation of products that promote tumorigenesis. In keeping with this hypothesis, we have found Mdm2-C localized to the nucleolus and the cytoplasm of T47D breast cancer cells (Okoro and Bargonetti, manuscript in preparation). T47D cells have mutant p53, and the translation of this mutant p53 might be promoted by Mdm2-C. The p53 mRNA is probably one of many mRNA target molecules for Mdm2-RNA interactions. This begs the question of why an Mdm2 isoform would increase p53 levels if its goal were to transform the cell. The answer to this might lie in the levels of other oncogenes or tumor suppressors expressed in the cell. The impact of Mdm2 isoforms could be profoundly influenced by their oligomerization relationship with Mdm2-FL and whether wild-type or mutant p53 is expressed in the cell. If Mdm2 isoforms increase the translation of gain-of-function mutant p53, they will potentiate tumorigenesis. Disrupting the Mdm2-RNA interaction by monoclonal antibodies to the C-terminus of Mdm258 would serve as an efficient approach to decreasing the effect of the mutant p53 protein and perhaps other oncogenic determinants.
Conclusion
Ultimately, the effect of the Mdm2 isoforms on cellular processes will depend on concentration and subcellular localization. The concentration of Mdm2 is influenced by a number of environmental factors. One example is the ability of estrogen to drive estrogen receptor positive cells to increase Mdm2 expression.65 Increases in estrogen concentration also turn on other signal transduction programs that activate cell growth. The concentration of Mdm2 is increased by promoter polymorphisms in the mdm2 gene sequence.63 In the presence of estrogen, the mdm2 SNP 309 genotype further drives expression of Mdm2 from the P2 promoter generating transcripts that will be efficiently translated.95,96 The concentration of Mdm2 isoforms may be driven by a cross-talk of environment and gene interactions.
Localization of Mdm2 isoforms to the nucleolus and cytoplasm may influence the RNA metabolism of many gene targets. It is highly likely that the targets of Mdm2 go far beyond p53, but they remain to be determined. When endogenous Mdm2 isoforms are detected with more regularity, we will need to stratify the localization patterns. The localization of Mdm2 isoforms in cancers will lend further evidence to propose functional consequences.
Acknowledgments
We thank members of the Bargonetti laboratory for critical comments on the manuscript. The National Science Foundation is acknowledged for supporting the Bargonetti laboratory as they work to elucidate the molecular determinants for Mdm2-mediated growth activation.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Dr. Bargonetti was funded by the National Science Foundation grant MCB-0744316 and The Breast Cancer Research Foundation. Danielle Okoro was partially supported by a CUNY Graduate Center Magnet Award. Melissa Rosso was partially supported by a MBRS-RISE fellowship 3R25- GM060665. Hunter College research is made possible in part by a Research Centers in Minority Institutions grant from the National Center for Research Resources (NCRR) (G12 RR003037). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR.
References
- 1. Haines DS, Landers JE, Engle LJ, George DL. Physical and functional interaction between wild-type p53 and mdm2 proteins. Mol Cell Biol. 1994;14:1171-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Finlay CA. The mdm-2 oncogene can overcome wild-type p53 suppression of transformed cell growth. Mol Cell Biol. 1993;13:301-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993-1000 [PubMed] [Google Scholar]
- 4. Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2:1-8 [PubMed] [Google Scholar]
- 5. Sigalas I, Calvert AH, Anderson JJ, Neal DE, Lunec J. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: transforming ability and frequent detection in human cancer. Nat Med. 1996;2:912-7 [DOI] [PubMed] [Google Scholar]
- 6. Matsumoto R, Tada M, Nozaki M, Zhang CL, Sawamura Y, Abe H. Short alternative splice transcripts of the mdm2 oncogene correlate to malignancy in human astrocytic neoplasms. Cancer Res. 1998;58:609-13 [PubMed] [Google Scholar]
- 7. Lukas J, Gao DQ, Keshmeshian M, et al. Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res. 2001;61:3212-9 [PubMed] [Google Scholar]
- 8. Liang H, Atkins H, Abdel-Fattah R, Jones SN, Lunec J. Genomic organisation of the human MDM2 oncogene and relationship to its alternatively spliced mRNAs. Gene. 2004;338:217-23 [DOI] [PubMed] [Google Scholar]
- 9. Sanchez-Aguilera A, Garcia JF, Sanchez-Beato M, Piris MA. Hodgkin’s lymphoma cells express alternatively spliced forms of HDM2 with multiple effects on cell cycle control. Oncogene. 2006;25:2565-74 [DOI] [PubMed] [Google Scholar]
- 10. Jeyaraj S, O’Brien DM, Chandler DS. MDM2 and MDM4 splicing: an integral part of the cancer spliceome. Front Biosci. 2009;14:2647-56 [DOI] [PubMed] [Google Scholar]
- 11. Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342-57 [DOI] [PubMed] [Google Scholar]
- 12. Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100-7 [DOI] [PubMed] [Google Scholar]
- 13. Bartel F, Taubert H, Harris LC. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell. 2002;2:9-15 [DOI] [PubMed] [Google Scholar]
- 14. Graveley BR. Splicing up pluripotency. Cell. 2011;147:22-4 [DOI] [PubMed] [Google Scholar]
- 15. Gabut M, Samavarchi-Tehrani P, Wang X, et al. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132-46 [DOI] [PubMed] [Google Scholar]
- 16. Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5-9 [DOI] [PubMed] [Google Scholar]
- 17. Coutts AS, Boulahbel H, Graham A, La Thangue NB. Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep. 2007;8:84-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei X, Yu ZK, Ramalingam A, et al. Physical and functional interactions between PML and MDM2. J Biol Chem. 2003;278:29288-97 [DOI] [PubMed] [Google Scholar]
- 19. Leveillard T, Wasylyk B. The MDM2 C-terminal region binds to TAFII250 and is required for MDM2 regulation of the cyclin A promoter. J Biol Chem. 1997;272:30651-61 [DOI] [PubMed] [Google Scholar]
- 20. Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23: 8902-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stevens C, Pettersson S, Wawrzynow B, et al. ATP stimulates MDM2-mediated inhibition of the DNA-binding function of E2F1. FEBS J. 2008;275:4875-86 [DOI] [PubMed] [Google Scholar]
- 23. Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci U S A. 2006; 103:1227-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14:7414-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sui G, Affar el B, Shi Y, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117: 859-72 [DOI] [PubMed] [Google Scholar]
- 26. Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/ neu-overexpressing cells. Nat Cell Biol. 2001;3: 245-52 [DOI] [PubMed] [Google Scholar]
- 27. Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998;95:8292-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grossman SR, Perez M, Kung AL, et al. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405-15 [DOI] [PubMed] [Google Scholar]
- 29. Leng P, Brown D, Deb S. Human oncoprotein mdm2 interacts with the tata-binding protein in-vitro and in-vivo. Int J Oncol. 1995;6:251-9 [PubMed] [Google Scholar]
- 30. Xiao ZX, Chen J, Levine AJ, et al. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694-8 [DOI] [PubMed] [Google Scholar]
- 31. Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem. 2005;280:18771-81 [DOI] [PubMed] [Google Scholar]
- 32. Johnson-Pais T, Degnin C, Thayer MJ. pRB induces Sp1 activity by relieving inhibition mediated by MDM2. Proc Natl Acad Sci U S A. 2001;98:2211-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thut CJ, Goodrich JA, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boyd MT, Vlatkovic N, Haines DS. A novel cellular protein (MTBP) binds to MDM2 and induces a G1 arrest that is suppressed by MDM2. J Biol Chem. 2000;275:31883-90 [DOI] [PubMed] [Google Scholar]
- 35. Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zeng X, Chen L, Jost CA, et al. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol. 1999;19:3257-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Calabro V, Mansueto G, Parisi T, Vivo M, Calogero RA, La Mantia G. The human MDM2 oncoprotein increases the transcriptional activity and the protein level of the p53 homolog p63. J Biol Chem. 2002;277:2674-81 [DOI] [PubMed] [Google Scholar]
- 38. Juven-Gershon T, Shifman O, Unger T, Elkeles A, Haupt Y, Oren M. The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol Cell Biol. 1998;18:3974-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin K, Trouche D, Hagemeier C, Sorensen TS, La Thangue NB, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375:691-4 [DOI] [PubMed] [Google Scholar]
- 40. Jung YS, Qian Y, Chen X. DNA polymerase eta is targeted by Mdm2 for polyubiquitination and proteasomal degradation in response to ultraviolet irradiation. DNA Repair (Amst). 2012;11:177-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vlatkovic N, Guerrera S, Li Y, Linn S, Haines DS, Boyd MT. MDM2 interacts with the C-terminus of the catalytic subunit of DNA polymerase epsilon. Nucleic Acids Res. 2000;28:3581-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wallace M, Worrall E, Pettersson S, Hupp TR, Ball KL. Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Mol Cell. 2006;23:251-63 [DOI] [PubMed] [Google Scholar]
- 43. Ma J, Martin JD, Zhang H, et al. A second p53 binding site in the central domain of Mdm2 is essential for p53 ubiquitination. Biochemistry. 2006;45:9238-45 [DOI] [PubMed] [Google Scholar]
- 44. Bennett-Lovsey R, Hart SE, Shirai H, Mizuguchi K. The SWIB and the MDM2 domains are homologous and share a common fold. Bioinformatics. 2002;18:626-30 [DOI] [PubMed] [Google Scholar]
- 45. Steinman HA, Burstein E, Lengner C, et al. An alternative splice form of Mdm2 induces p53-independent cell growth and tumorigenesis. J Biol Chem. 2004;279:4877-86 [DOI] [PubMed] [Google Scholar]
- 46. Volk EL, Fan L, Schuster K, Rehg JE, Harris LC. The MDM2-a splice variant of MDM2 alters transformation in vitro and the tumor spectrum in both Arf- and p53-null models of tumorigenesis. Mol Cancer Res. 2009;7:863-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bartel F, Meye A, Wurl P, et al. Amplification of the MDM2 gene, but not expression of splice variants of MDM2 MRNA, is associated with prognosis in soft tissue sarcoma. Int J Cancer. 2001;95:168-75 [DOI] [PubMed] [Google Scholar]
- 48. Bartel F, Harris LC, Wurl P, Taubert H. MDM2 and its splice variant messenger RNAs: expression in tumors and down-regulation using antisense oligonucleotides. Mol Cancer Res. 2004;2:29-35 [PubMed] [Google Scholar]
- 49. Taubert H, Bartel F, Greither T, et al. Association of HDM2 transcript levels with age of onset and prognosis in soft tissue sarcomas. Mol Cancer Res. 2008;6:1575-81 [DOI] [PubMed] [Google Scholar]
- 50. Pinkas J, Naber SP, Butel JS, Medina D, Jerry DJ. Expression of MDM2 during mammary tumorigenesis. Int J Cancer. 1999;81:292-8 [DOI] [PubMed] [Google Scholar]
- 51. Brekman A, Singh KE, Polotskaia A, Kundu N, Bargonetti J. A p53-independent role of Mdm2 in estrogen-mediated activation of breast cancer cell proliferation. Breast Cancer Res. 2011;13:R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci U S A. 1998;95:15608-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fridman JS, Hernando E, Hemann MT, de Stanchina E, Cordon-Cardo C, Lowe SW. Tumor promotion by Mdm2 splice variants unable to bind p53. Cancer Res. 2003;63:5703-6 [PubMed] [Google Scholar]
- 54. Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol. 2000;2:179-81 [DOI] [PubMed] [Google Scholar]
- 55. Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine AJ. The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol Med. 1996;2:439-51 [PMC free article] [PubMed] [Google Scholar]
- 56. Wawrzynow B, Zylicz A, Wallace M, Hupp T, Zylicz M. MDM2 chaperones the p53 tumor suppressor. J Biol Chem. 2007;282:32603-12 [DOI] [PubMed] [Google Scholar]
- 57. Challen C, Anderson JJ, Chrzanowska-Lightowlers ZM, Lightowlers RN, Lunec J. Recombinant human MDM2 oncoprotein shows sequence composition selectivity for binding to both RNA and DNA. Int J Oncol. 2012;40:851-9 [DOI] [PubMed] [Google Scholar]
- 58. Anderson JJ, Challen C, Atkins H, Suaeyun R, Crosier S, Lunec J. MDM2 RNA binding is blocked by novel monoclonal antibody h-MDM2-F4-14. Int J Oncol. 2007;31:545-55 [PubMed] [Google Scholar]
- 59. Yu GW, Allen MD, Andreeva A, Fersht AR, Bycroft M. Solution structure of the C4 zinc finger domain of HDM2. Protein Sci. 2006;15:384-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Naski N, Gajjar M, Bourougaa K, Malbert-Colas L, Fahraeus R, Candeias MM. The p53 mRNA-Mdm2 interaction. Cell Cycle. 2009;8:31-4 [DOI] [PubMed] [Google Scholar]
- 61. Chandler DS, Singh RK, Caldwell LC, Bitler JL, Lozano G. Genotoxic stress induces coordinately regulated alternative splicing of the p53 modulators MDM2 and MDM4. Cancer Res. 2006;66:9502-8 [DOI] [PubMed] [Google Scholar]
- 62. Dias CS, Liu Y, Yau A, Westrick L, Evans SC. Regulation of hdm2 by stress-induced hdm2alt1 in tumor and nontumorigenic cell lines correlating with p53 stability. Cancer Res. 2006;66:9467-73 [DOI] [PubMed] [Google Scholar]
- 63. Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591-602 [DOI] [PubMed] [Google Scholar]
- 64. Saucedo LJ, Myers CD, Perry ME. Multiple murine double minute gene 2 (MDM2) proteins are induced by ultraviolet light. J Biol Chem. 1999;274:8161-8 [DOI] [PubMed] [Google Scholar]
- 65. Bond GL, Hirshfield KM, Kirchhoff T, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104-10 [DOI] [PubMed] [Google Scholar]
- 66. Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23:2584-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994;8:1739-49 [DOI] [PubMed] [Google Scholar]
- 68. Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G. An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene. 2001;20:4041-9 [DOI] [PubMed] [Google Scholar]
- 69. Volk EL, Schuster K, Nemeth KM, Fan L, Harris LC. MDM2-A, a common Mdm2 splice variant, causes perinatal lethality, reduced longevity and enhanced senescence. Dis Model Mech. 2009;2:47-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Olson DC, Marechal V, Momand J, Chen J, Romocki C, Levine AJ. Identification and characterization of multiple mdm-2 proteins and mdm-2-p53 protein complexes. Oncogene. 1993;8:2353-60 [PubMed] [Google Scholar]
- 71. Schuster K, Fan L, Harris LC. MDM2 splice variants predominantly localize to the nucleoplasm mediated by a COOH-terminal nuclear localization signal. Mol Cancer Res. 2007;5:403-12 [DOI] [PubMed] [Google Scholar]
- 72. Dang J, Kuo ML, Eischen CM, Stepanova L, Sherr CJ, Roussel MF. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 2002;62:1222-30 [PubMed] [Google Scholar]
- 73. Poyurovsky MV, Prives C. Unleashing the power of p53: lessons from mice and men. Genes Dev. 2006;20:125-31 [DOI] [PubMed] [Google Scholar]
- 74. Ganguli G, Wasylyk B. p53-independent functions of MDM2. Mol Cancer Res. 2003;1:1027-35 [PubMed] [Google Scholar]
- 75. Bouska A, Eischen CM. Mdm2 affects genome stability independent of p53. Cancer Res. 2009;69:1697-701 [DOI] [PubMed] [Google Scholar]
- 76. Tovar C, Rosinski J, Filipovic Z, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Marechal V, Elenbaas B, Taneyhill L, et al. Conservation of structural domains and biochemical activities of the MDM2 protein from Xenopus laevis. Oncogene. 1997;14:1427-33 [DOI] [PubMed] [Google Scholar]
- 78. Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296-9 [DOI] [PubMed] [Google Scholar]
- 79. Deb SP. Cell cycle regulatory functions of the human oncoprotein MDM2. Mol Cancer Res. 2003;1:1009-16 [PubMed] [Google Scholar]
- 80. Poyurovsky MV, Katz C, Laptenko O, et al. The C terminus of p53 binds the N-terminal domain of MDM2. Nat Struct Mol Biol. 2010;17:982-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Meek DW, Knippschild U. Posttranslational modification of MDM2. Mol Cancer Res. 2003;1:1017-26 [PubMed] [Google Scholar]
- 82. Kraus A, Neff F, Behn M, Schuermann M, Muenkel K, Schlegel J. Expression of alternatively spliced mdm2 transcripts correlates with stabilized wild-type p53 protein in human glioblastoma cells. Int J Cancer. 1999;80:930-4 [DOI] [PubMed] [Google Scholar]
- 83. Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat Chem Biol. 2009;5:778-81 [DOI] [PubMed] [Google Scholar]
- 84. Tsvetkov P, Reuven N, Prives C, Shaul Y. Susceptibility of p53 unstructured N terminus to 20 S proteasomal degradation programs the stress response. J Biol Chem. 2009;284:26234-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nie L, Sasaki M, Maki CG. Regulation of p53 nuclear export through sequential changes in conformation and ubiquitination. J Biol Chem. 2007;282:14616-25 [DOI] [PubMed] [Google Scholar]
- 86. Lushnikova T, Bouska A, Odvody J, Dupont WD, Eischen CM. Aging mice have increased chromosome instability that is exacerbated by elevated Mdm2 expression. Oncogene. 2011;30:4622-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene 2008;27:1590-8 [DOI] [PubMed] [Google Scholar]
- 88. Priest C, Prives C, Poyurovsky MV. Deconstructing nucleotide binding activity of the Mdm2 RING domain. Nucleic Acids Res. 2010;38:7587-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631-9 [DOI] [PubMed] [Google Scholar]
- 90. Arva NC, Talbott KE, Okoro DR, Brekman A, Qiu WG, Bargonetti J. Disruption of the p53-Mdm2 complex by Nutlin-3 reveals different cancer cell phenotypes. Ethn Dis. 2008;18:S2-1-8 [PMC free article] [PubMed] [Google Scholar]
- 91. Arva NC, Gopen TR, Talbott KE, et al. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J Biol Chem. 2005;280: 26776-87 [DOI] [PubMed] [Google Scholar]
- 92. Gajjar M, Candeias MM, Malbert-Colas L, et al. The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell. 2012;21:25-35 [DOI] [PubMed] [Google Scholar]
- 93. Candeias MM, Malbert-Colas L, Powell DJ, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10:1098-105 [DOI] [PubMed] [Google Scholar]
- 94. Hamard PJ, Manfredi JJ. Mdm2’s dilemma: to degrade or to translate p53? Cancer Cell. 2012;21:3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Landers JE, Haines DS, Strauss JF, III, George DL. Enhanced translation: a novel mechanism of mdm2 oncogene overexpression identified in human tumor cells. Oncogene. 1994;9:2745-50 [PubMed] [Google Scholar]
- 96. Landers JE, Cassel SL, George DL. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res. 1997;57: 3562-8 [PubMed] [Google Scholar]