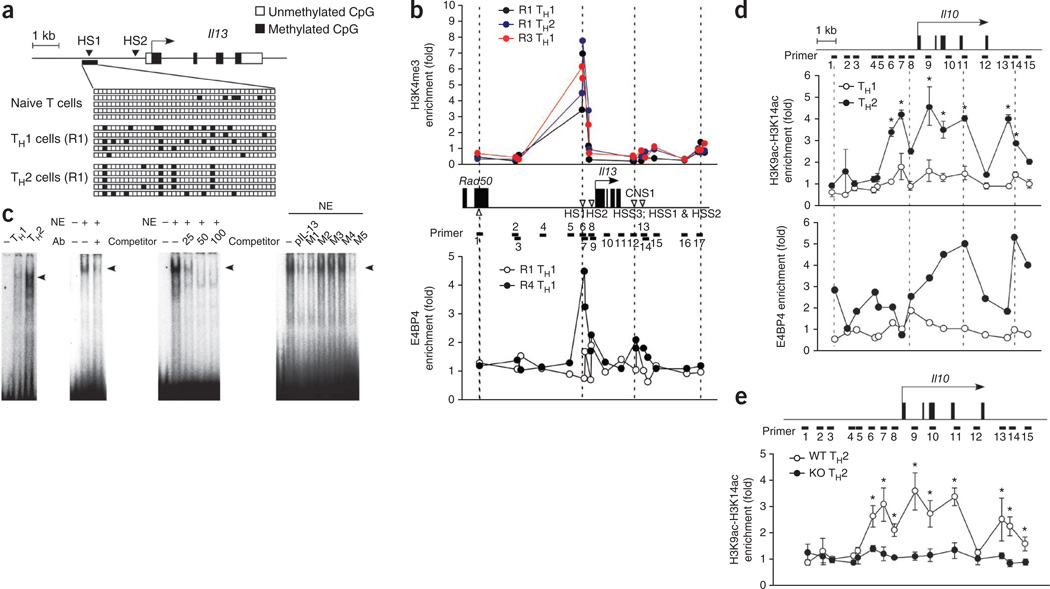
Figure 7.
E4BP4 protein binds specifically to the Il13 promoter and in the Il10 locus. (a) Methylation of genomic DNA (bottom) isolated from naive CD4+ T cells and from TH1 cells and TH2 cells given 1 week of stimulation (R1), analyzed by sequencing with primers specific for the Il13 promoter (top) after bisulfite treatment. (b) ChIP analysis of the enrichment of histone H3 trimethylated at Lys4 (H3K4me3) in R1 TH1 cells, R1 TH2 cells or R3 TH1 cells (top) and of E4BP4 in R1 TH1 and R4 TH1 cells (bottom), both at the Il13 locus (middle). CNS1, conserved noncoding sequence 1; HSS, hypersensitive site; Rad50, gene encoding a protein involved in the repair of DNA double-strand breaks. (c) Electrophoretic mobility-shift assay of nuclear extracts (NE) of TH1 and TH2 cells, with an oligonucleotide from a specific region of the Il13 promoter as the probe (far left); supershift analysis of nuclear extracts from TH2 cells with anti-E4BP4 (Ab; middle left); and competition analysis with 25-, 50- or 100-fold excess competitor (middle right) or mutant oligonucleotides (M1–M5) and the Il13 promoter (pIL-13; far right). (d) ChIP analysis of the enrichment of histone H3 acetylated at Lys9 and Lys14 (H3K9ac–H3K14ac; top) or of E4BP4 (bottom) at the Il10 locus (above) in chromatin fractions extracted from TH1 or TH2 cells; results are presented relative to those obtained with input DNA prepared from untreated chromatin. (e) ChIP analysis of the enrichment of histone H3 acetylated at Lys9 and Lys14 at the Il10 locus (above) in TH2 cells from E4bp4−/− and wild-type mice; results presented as in d. *P < 0.01 (Student’s t-test). Data are representative of two experiments (a–c) or are from three independent experiment (d,e; mean and s.e.m.).