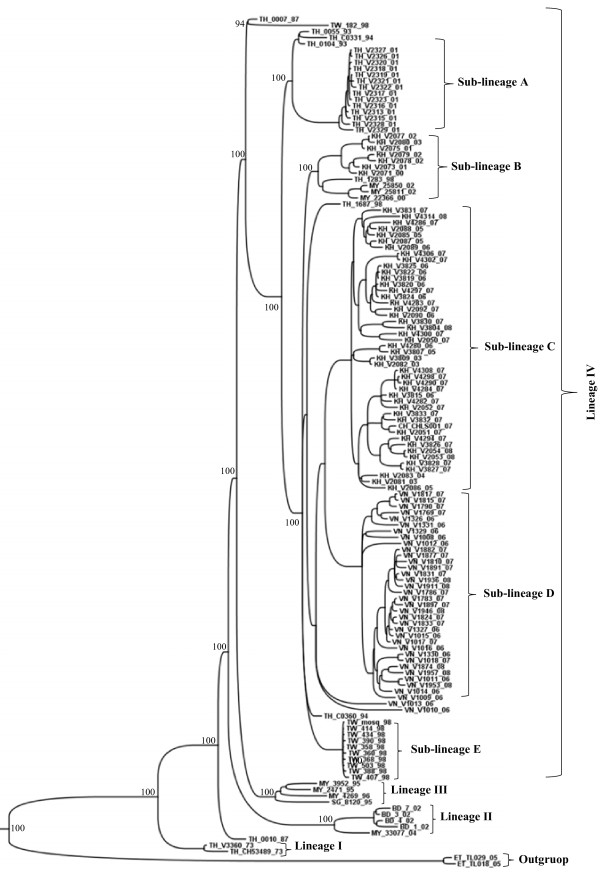
Figure 6.
Bayesian phylogenetic trees based on the entire ORFs derived from 137 global samples of the genotype II DENV-3 inferred with MrBayes program. The posterior probabilities are expressed in percent and indicated at important nodes. The best fit-model of nucleotide substitution for Bayesian phylogenetic reconstruction used was under a General Time Reversible model with gamma-distributed rate variation (G = 1.4745) and a proportion of invariable sites (I = 0.4956) (GTR + G + I). Branch lengths are proportional to percentage of divergence.