Table 2. Summary of the methods.
| Simes test | Fisher test | GATES | VEGAS | FOSCO | |
| Source | [12] | [11] | [10] | [9] | – |
| Core idea | Adjust p-values by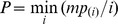 under assumption of independence under assumption of independence |
Obtain p-value based on 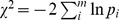 under the assumption of independence under the assumption of independence |
Adjust p-values by 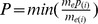 , where me(i) is obtained from each gene empirically , where me(i) is obtained from each gene empirically |
Obtain p-values base on 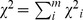 . The distribution under the null hypothesis is obtained through Monte Carlo methods
. The distribution under the null hypothesis is obtained through Monte Carlo methods |
Adjust p-values by 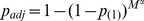 where α is determined empirically from a random permutation where α is determined empirically from a random permutation |
| Linear regression coefficient after correction* | 2.32E-04 | −8.32E-03 | −3.14E-04 | −1.00E-04 | −3.75E-04 |
| SNPs association to genes | NCBI dbSNPs | NCBI dbSNPs | NCBI dbSNPs+5kb flanking regions | UCSG Genome Brower hg18+50 kb flanking regions | NCBI dbSNPs |
Note: * Linear regression coefficient before correction β = −7.23E-03.
