Figure 1.
Genome-wide Analysis of Sen1 and Nrd1 Localization during the Cell Cycle
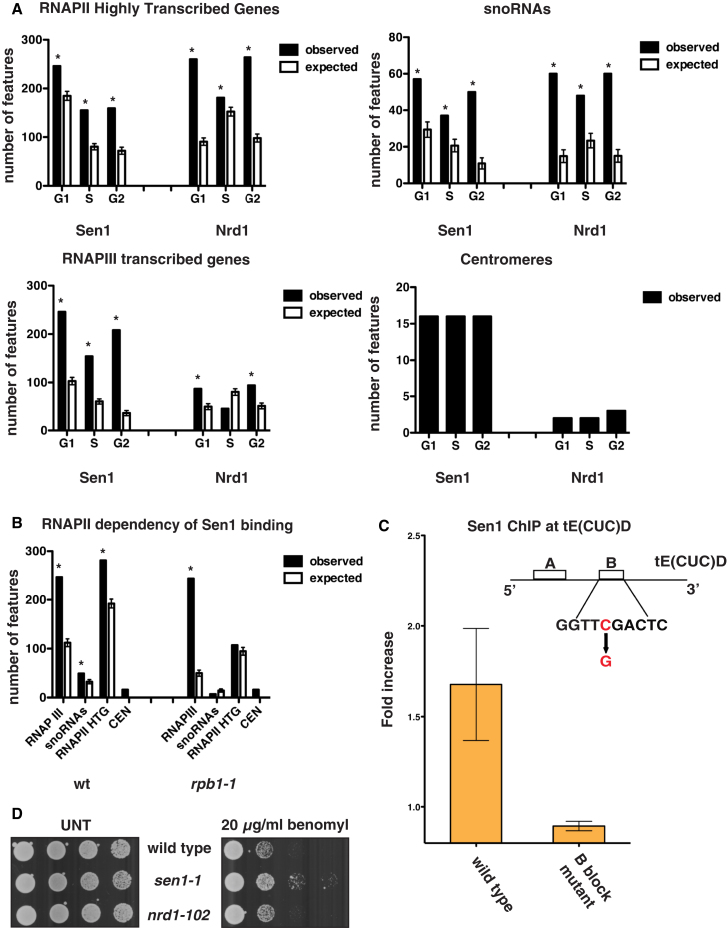
(A) SEN1- (Sen1) and NRD1-FLAG (Nrd1) strains were blocked in G1 phase by α-factor treatment (G1) and were released at 23°C in either 0.2 M HU for 60 min (S phase; S) or nocodazole for 180 min (G2/M). Samples were collected in each cell-cycle phase to analyze the genomic distribution of factors by ChIP-chip. The number of features bound by each factor in the ChIP experiments (observed) as well as in bioinformatic simulations is shown for highly transcribed RNAPII genes, snoRNAs, RNAPIII genes, and centromeres (see Experimental Procedures). Data represent the mean ±SD of three independent simulations (expected). Asterisks indicate features showing a statistically significant difference between observed and simulated binding. The statistical significance of Sen1 and Nrd1 binding to centromeres could not be calculated by this method due to the reduced total number—sixteen—of these elements.
(B) WT and rpb1-1 cells were arrested in G2/M phase as in Figure 1A, transferred to 37°C for 1 hr to inactivate the RNAPII, and then processed for ChIP-chip to analyze Sen1 binding.
(C) Sen1 binding to a WT or a B-block promoter mutant tE(CUC)D gene was evaluated by ChIP, followed by qPCR in G2/M-arrested cells. Data represent the mean ±SD of three independent experiments.
(D) Serial dilutions of WT, sen1-1, and nrd1-102 cells plated in the presence or absence (UNT) of benomyl.
See also Figure S1.