Abstract
Huntington's disease (HD) is a dominantly inherited neurodegenerative disorder caused by expanded CAG repeats in the huntingtin (HTT) gene. Although several palliative treatments are available, there is currently no cure and patients generally die 10–15 y after diagnosis. Several promising approaches for HD therapy are currently in development, including RNAi and antisense analogs. We developed a complementary strategy to test repression of mutant HTT with zinc finger proteins (ZFPs) in an HD model. We tested a “molecular tape measure” approach, using long artificial ZFP chains, designed to bind longer CAG repeats more strongly than shorter repeats. After optimization, stable ZFP expression in a model HD cell line reduced chromosomal expression of the mutant gene at both the protein and mRNA levels (95% and 78% reduction, respectively). This was achieved chromosomally in the context of endogenous mouse HTT genes, with variable CAG-repeat lengths. Shorter wild-type alleles, other genomic CAG-repeat genes, and neighboring genes were unaffected. In vivo, striatal adeno-associated virus viral delivery in R6/2 mice was efficient and revealed dose-dependent repression of mutant HTT in the brain (up to 60%). Furthermore, zinc finger repression was tested at several levels, resulting in protein aggregate reduction, reduced decline in rotarod performance, and alleviation of clasping in R6/2 mice, establishing a proof-of-principle for synthetic transcription factor repressors in the brain.
Keywords: gene therapy, transcription repression, protein engineering, protein design, synthetic biology
The pathological expansion of CAG repeats leads to extended polyglutamine (polyQ) tracts in mutated gene products (1). The resulting proteins are thought to aggregate and cause toxic gain-of-function diseases, including spinocerebellar ataxias, spinobulbar muscular atrophy, and Huntington's disease (HD) (2, 3). HD neuropathology is associated with selective neuronal cell death, primarily of medium spiny neurons of the striatum and, to a lesser extent, cortical neurons, causing cognitive dysfunction and chorea (1, 4). Since the discovery, in 1993, that the huntingtin (HTT) gene caused HD (5), much attention has been focused on how the CAG-repeat number affects the pathology and progression of the disease; normally, the number of CAG repeats in the wild-type (wt) HTT gene ranges from 10 to 29 (median of 18) (1, 4), whereas that of patients with HD typically ranges from 36 to 121 (median of 44) (1, 4). Because the age of onset of disease is correlated to CAG-repeat number (1), reducing the polyQ “load” on cells should be beneficial therapeutically.
Despite intense research, there is no way to stop or delay the progression of HD and current treatments merely treat symptoms (1, 4). However, a number of promising studies have aimed at improving cell survival of affected areas (reviewed in 6). Unlike other neurological disorders, such as Alzheimer’s disease and Parkinson disease, HD is monogenic (5). Therefore, therapeutic strategies need only target the expression of the causal gene to reverse and treat the effects of the mutant protein. However, because wt HTT protein is widely expressed (7), is essential for early embryonic development (8), and is required for neuronal function and survival in the brain (9), it is important to reduce the expression of the mutant protein specifically, while leaving the expression of the wt protein as unaffected as possible.
Several approaches using synthetic nucleic acids that selectively target the mutant allele are currently being developed (reviewed in 10). Recently, RNAi was shown to reduce expression of mutant HTT (11–13). Although this technique could be very powerful, mutant-selective RNAi depends on targeting single nucleotide or deletion polymorphisms that differentiate between alleles, and these often differ from patient to patient. However, there is evidence that partial repression of wt HTT can be tolerated (14, 15), suggesting that generic approaches that repress both alleles should also be pursued. Peptide nucleic acids and locked nucleic acids are generic; yet, some promising partially selective inhibition of expanded CAG repeats of the ataxin-3 and HTT genes has been reported (16, 17). Recently, there has even been sustained disease reversal using antisense oligonucleotides (18).
Also aiming to reduce the levels of mutant protein, a different approach was proposed by Bauer et al. (19), who designed a polyQ-binding peptide, fused to heat shock cognate protein 70 binding motif, to direct degradation of mutant HTT via chaperone-mediated autophagy. Intrastriatal recombinant adeno-associated virus (rAAV) delivery of this fusion protein showed a strong therapeutic effect.
In this study, we examined the use of zinc finger proteins (ZFPs) as a complementary approach to reduce the expression of mutant HTT, which could be used in tandem with any of the above approaches. Because zinc fingers can be readily reengineered to bind different DNA sequences (20–29), including CAG repeats (30), they could, in principle, be used to target the HTT gene at a transcriptional level. Furthermore, zinc fingers can be easily concatenated into long chains, and different linker designs can alter the interaction kinetics substantially (31). This suggested to us that a systematic appraisal of different-length ZFP, with appropriate linker designs, might reveal an optimal configuration for repressing mutant HTT, while leaving the wt allele relatively unaffected.
We therefore designed a ZFP able to recognize and bind poly-5′-GC (A/T)-3′, such that it would recognize both poly-CAG and its complementary DNA strand. The resulting ZFP chains (4–18 fingers) were able to repress target genes with longer CAG repeats preferentially, compared with shorter repeats. This was carried out both in transient transfection reporter assays and with stable expression for 20 d, against full HTT chromosomal targets, in various model cell lines for HD. Ultimately, we tested the most effective ZFP in vivo in the R6/2 HD mouse model, using AAV for delivery, establishing a proof-of-principle for gene repression by synthetic zinc fingers in the brain.
Results
Designing Long ZFP Chains to Bind Expanded CAG Repeats.
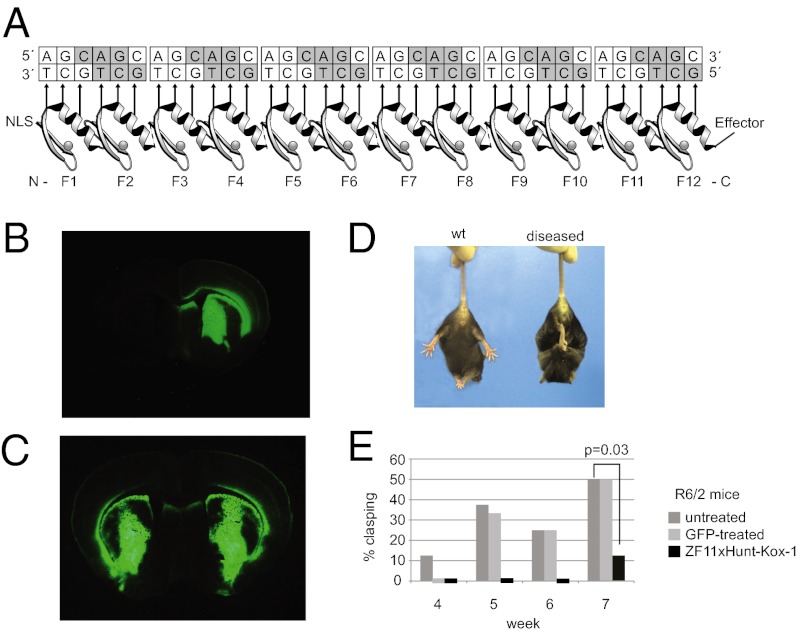
Zinc fingers can be concatenated to make long multifinger chains (31, 32), and, to date, no one has systematically explored the binding modes of different-length ZFP to long repetitive DNA tracts. We therefore used rational design to construct a zinc finger (ZFxHunt) that would bind 5′-GC (A/T)-3′, such that polyfinger proteins would bind poly-GCA and poly-GCT (Materials and Methods and Fig. 1). Both DNA strands were targeted to increase the avidity for low-copy chromosomal targets. For structural reasons, zinc finger chains with canonical linkers lose their register with cognate DNA after three fingers. Therefore, extra -Gly (31) or 29-residue linkers were added after every two and six fingers, respectively (full sequences are given in Dataset S1). In this way, different numbers of fingers could be tested for length-dependent discrimination.
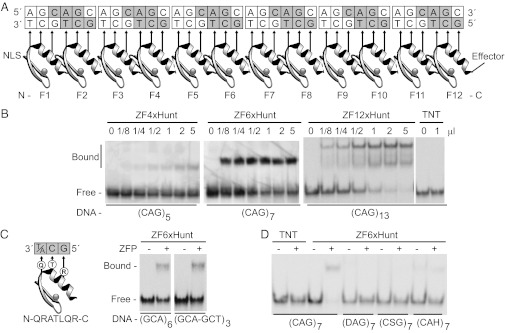
Fig. 1.
Zinc finger arrays to bind CAG repeats. (A) A twelve-finger array shows recognition helices contacting 5′-GCT-3′ bases on the lower DNA strand. Similar arrays of 4, 6, 12, or 18 zinc fingers were built (ZF4xHunt, ZF6xHunt, ZF12xHunt, and ZF18xHunt). Nuclear localization signals (NLS) and effectors (e.g., Kox-1 transcription repression domain) were added to N and C termini, respectively. (B) Gel shift assays show 4-, 6-, or 12-finger arrays binding poly-CAG DNA and forming distinct complexes. TNT is a negative control. (C) A hybrid zinc finger design recognizes 5′-GC(A/T)-3′, allowing binding to either the (GCA)n or (GCT)n complementary strand of the CAG repeat. A gel shift assay shows equal binding to GCA or GCT triplets in mixed sequences. (D) Specificity gel shift assay. Zinc fingers bind preferentially to CAG repeats (CAG7) compared with degenerate mutant sequences (DAG7, CSG7, or CAH7; D = A, G, T; S = C, G; H = A, C, T).
First, zinc finger chains containing either 4, 6, or 12 ZFxHunt domains were constructed and tested in gel shift assays for binding to double-stranded CAG probes; longer ZFPs gave more complete binding (Fig. 1B). Intriguingly, distinct bound complexes were observed, indicating that the ZFPs found single thermodynamic equilibria and were not trapped by kinetic intermediates. Highly repetitive zinc fingers and DNA might have been expected to form contiguous partial binding events, which would have resulted in broad smears in gel shifts, but this is not the case. Nonetheless, the 12-finger ZFP did give a lower secondary shift, presumably caused by a 6-finger degradation byproduct [zinc fingers can be unstable in linker regions (33)]. To our knowledge, these 12-finger (and subsequently 18-finger) domains are the longest functional artificial ZFP chains ever built.
To test whether ZFxHunt proteins are able to bind both strands of a CAG DNA probe, ZF6xHunt was assayed by gel shift and was shown to bind equally to both a CAG repetitive probe, (GCA)x6, and to an alternate CAG-CTG probe, (GCA-GCT)x3 (Fig. 1C). Furthermore, compared with mutated sequences, ZF6xHunt showed specificity for (CAG)x7 (Fig. 1D). In summary, we built multifinger ZFP able to bind poly 5′-GC (A/T)-3′ DNA probes specifically and efficiently in vitro.
Repression of PolyQ Reporter Genes.
Our starting hypothesis was that zinc finger proteins could preferentially bind longer CAG repeats through a mass action mechanism. Longer CAG repeats contain more target sites, and so should be bound and repressed more at any given ZFP concentration. Similarly, longer ZFP chains should also have a higher affinity, allowing one to balance expression, repression, and length preference. Although plasmid-based assays ultimately have different mass action kinetics from single-copy target chromosomal assays, they allowed us to verify our hypothesis rapidly before moving on to chromosomal targets.
ZFPs of different lengths were therefore tested using reporter vectors with N-terminal CAG repeats from the human HTT gene in frame with EGFP (Q0, Q10, Q35, and Q104; Q = CAG). To assess nonspecific effects, an HcRed reporter was cloned in a different region of the same vector under an independent promoter (Fig. 2A). HEK293T cells were transiently cotransfected with the indicated reporter and ZFxHunt vectors, driven by CMV promoters. Because gene therapy delivery vectors (used later) have a limited packaging capacity, which is exceeded by 12 fingers, we also tested an 11-finger construct. We found a similar expression level and activity between ZF12xHunt and ZF11xHunt (Fig. S1), and so used ZF11xHunt in all subsequent assays.
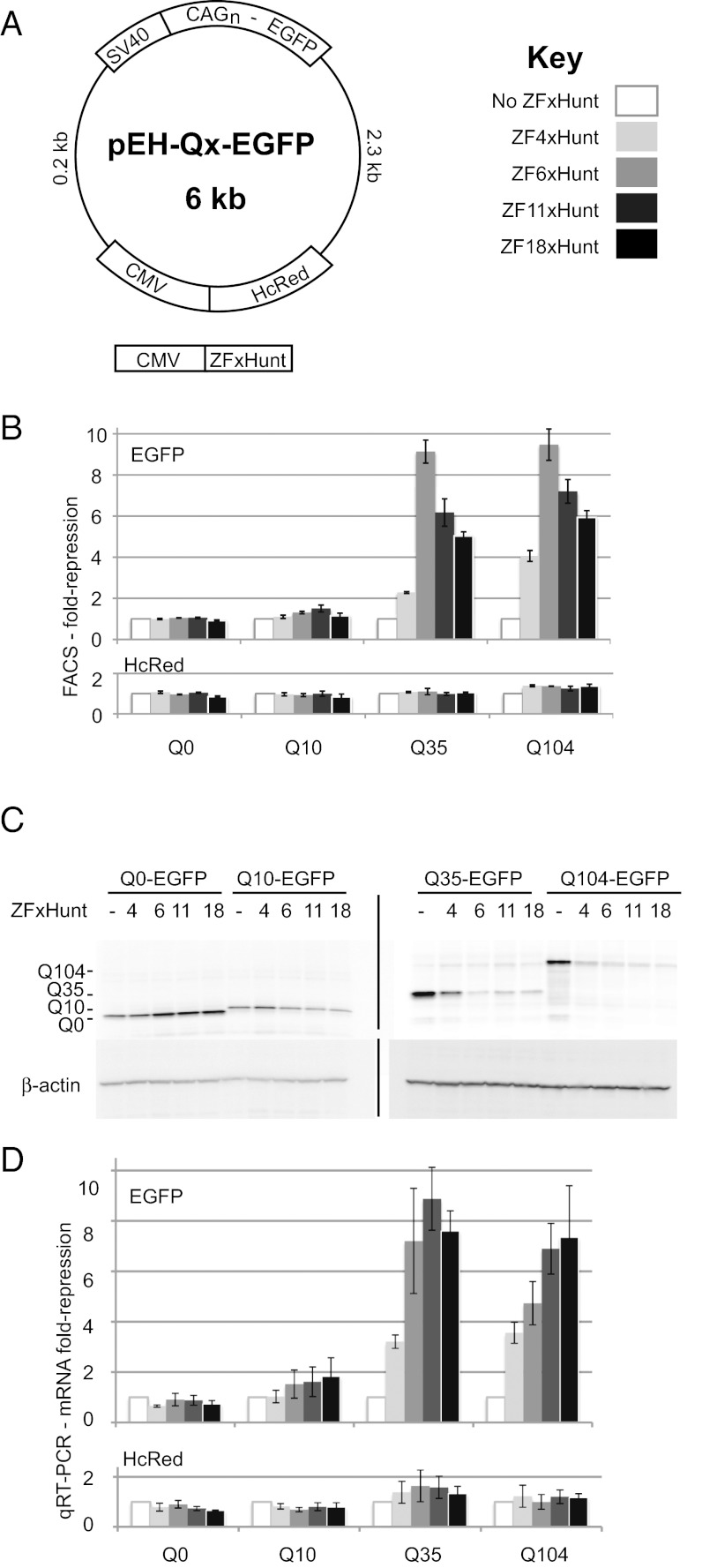
Fig. 2.
Episomal poly-CAG reporter repression by ZFxHunt with 0–18 fingers. (A) pEH reporter plasmids contain EGFP, fused to the N-terminal CAG repeats of the human HTT gene, expressing different-length polyQ coding sequences under an SV40 promoter. A control HcRed gene, under a CMV promoter, measures off-target repression. (B) FACS assay to measure the fold reduction in EGFP and HcRed fluorescent cells, in response to different zinc fingers. A 10-fold repression is equivalent to a 90% reduction. Results are the mean ± SEM of three independent experiments. (C) EGFP Western blot for ZFP repression of pEH-Qx targets. (D) qRT-PCR assay to measure fold repression of EGFP or HcRed mRNA by ZFP. Results are the mean ± SEM of four independent experiments.
Three assays were used to measure ZFxHunt repression: quantifying EGFP and HcRed fluorescent cells using FACS, EGFP protein levels in Western blots, and EGFP and HcRed mRNA levels in quantitative RT-PCR (qRT-PCR) (Fig. 2 B–D). In these experiments, ZFxHunt with 6, 11, and 18 fingers all repressed the two longer CAG repeats strongly (fivefold to ninefold, 80–90% repression), whereas ZF4xHunt was slightly weaker (twofold to fourfold, 50–75% repression). A Student’s t test analysis revealed that ZF11xHunt gave significant repression of longer polyQ reporter genes the most consistently of all zinc finger constructs, with no significant unspecific repression of the HcRed controls (P values in Tables S1 and S2). Verifying that the results were not specific to ZFP under the CMV promoter, similar results were obtained with the phosphoglycerate kinase promoter (Fig. S2). Although binding interactions (and thus particular length preference) can always be modulated by changing the effective concentration of each species, the main conclusion that can be drawn here is that there is a positive relationship between CAG repeat number and repression level. Furthermore, ZF6xHunt, ZF11xHunt, and ZF18xHunt are all strong repressors of longer repeats under these conditions.
Competition Assays Show Preferential Repression of Long CAG Repeats by ZFxHunt.
Up to this point, the zinc fingers had displayed length preference for single CAG-repeat targets in cells. Another test of potential length preference is for discrimination against two targets per cell, each with a different number of CAG repeats. HEK293T cells were therefore cotransfected with three plasmids: the indicated polyQ-EGFP and polyQ-mCherry reporter vectors, together with various ZFxHunt vectors (Fig. 3). The relative expression of the two reporters was measured by FACS (EGFP-positive or mCherry-positive cells). Fig. 3 shows that the longer CAG repeats are preferentially repressed by all ZFxHunt, such that cells are dominated by the shorter green or red constructs when their opposite counterpart is longer; this is seen directly by looking at the ratio of green to red expression. This can be explained most simply by mass action (longer CAG repeats contain more target sites, and thus are bound and repressed more).
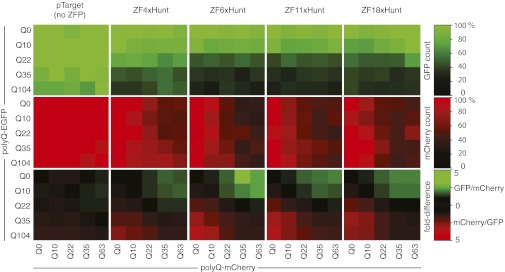
Fig. 3.
ZFP competition assay against pairs of different-length CAG repeats. Each small square represents one transfection experiment, where cells simultaneously receive two reporter plasmids, polyQ-EGFP and polyQ-mCherry, of different lengths (Q0 to Q104). ZFxHunt constructs with 4, 6, 11, or 18 fingers were tested for their ability to reduce the number of detectable green and red cells in FACS assays (%). Longer polyQ constructs are repressed preferentially, resulting in the shorter green or shorter red polyQ constructs dominating expression.
This assay demonstrates the potential of ZFxHunt for CAG-length discrimination. Therefore, length preference exists, even for multiple different-length CAG repeats within the same cell. The remaining caveat is that the system still needs to be tailored (expression levels, zinc finger length, and repression strength) for the particular cellular environment. We therefore moved next to testing in a more physiological chromosomal cellular environment.
Chromosomal Repression of Mutant HTT in an HD Model Cell Line.
Because ZF4xHunt had weaker binding and length preference (Figs. 2 and 3), and because ZF12xHunt and ZF18xHunt are beyond the packaging limit of AAV, we focused on testing ZF6xHunt and ZF11xHunt on chromosomal HTT genes. STHdh cells (34) are an established neuronal progenitor cell line from embryonic day 14 striatal primordia, derived from wt mice (STHdhQ7/HdhQ7) or knock-ins, where the first exon of the mouse HTT gene has been replaced by a human exon with 111 CAG repeats (STHdhQ111/HdhQ111 or STHdhQ7/HdhQ111). Because the chromosomal targets are present at a much lower effective concentration, to increase the repression per binding event, we also tested an effector domain in parallel: the Kox-1 KRAB domain (35).
STHdh cells stably expressing “naked” or Kox-1–fused ZF6xHunt and ZF11xHunt were harvested 20 d after retroviral infection, and huntingtin levels were analyzed by Western blot and qRT-PCR (Fig. 4A). Neither protein nor RNA levels of wt HTT (Q7) were reduced by naked or Kox-1–fused ZF6xHunt and ZF11xHunt. By contrast, Q111-mutant HTT RNA and protein levels were repressed with ZF6xHunt–Kox-1 by up to 2.5-fold (60% reduction) and twofold (50% reduction), respectively. Similar results were achieved in Q7/Q111 heterozygous cells (Fig. S3). ZF11xHunt–Kox-1 showed even stronger repression, with almost 80% reduction in mRNA expression and 95% reduction in the protein levels. Overall, the longer 11-finger construct, together with stronger Kox-1 repression, displayed the strongest chromosomal repression of mutant HTT.
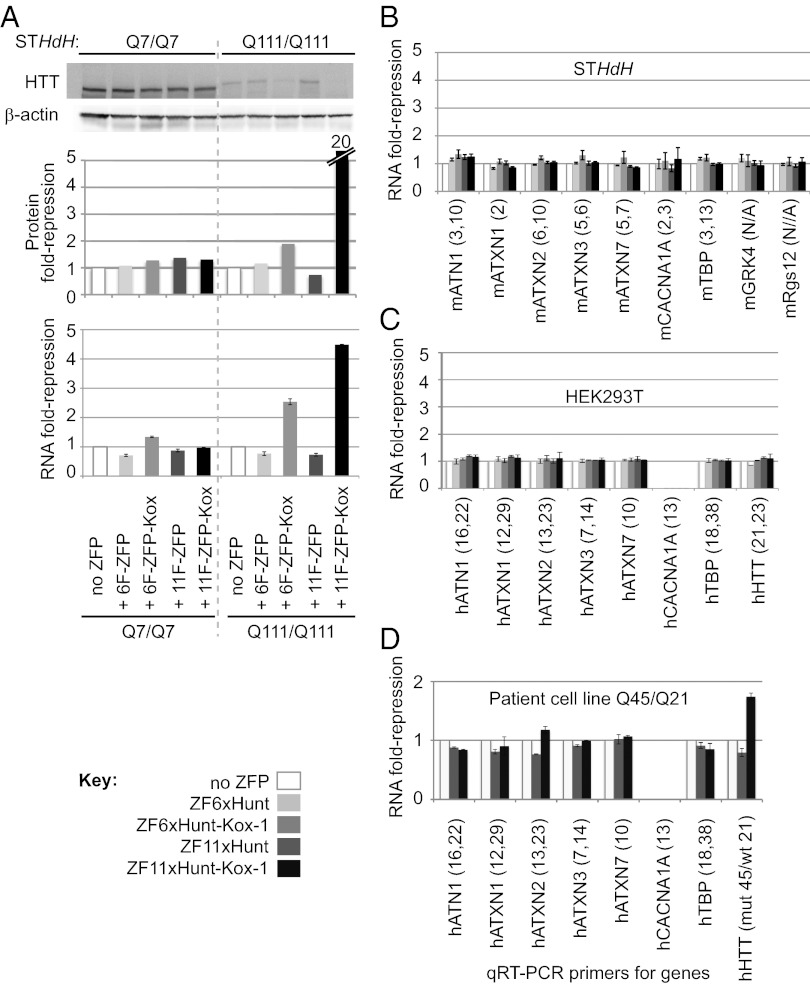
Fig. 4.
Expression of chromosomal CAG-repeat genes 20 d after retroviral ZFP delivery. Assays were carried out in wt mouse STHdh cells (Q7/Q7), in polyQ STHdh mutants (Q111/Q111), and in human HEK293T cells, as indicated. Mouse and human genes are prefixed by “m” and “h,” respectively. (A) Repression of endogenous HTT by ZF6xHunt and ZF11xHunt, with and without Kox-1 repressor domain. Western blots for HTT (Top) were controlled with β-actin staining and quantified using ImageJ (protein fold repression; Middle). qRT-PCR was used to compare HTT mRNA levels (RNA fold repression; Bottom). The experiment was repeated independently three times with similar results; one experiment is displayed. (B) mRNA levels of other wt CAG-repeat genes are broadly unaffected in STHdh cells (pooled samples: 3 Q7/Q7 and 3 Q111/Q111). Seven genes were tested by qRT-PCR (ATN1, atrophin1; ATXN1–3, 7, ataxin-1–3, 7; CACNA1A, calcium channel α1A subunit; TBP, TATA binding protein). CAG-repeat numbers are in brackets; the first number corresponds to pure CAG repeats, and the second number corresponds to broken CAG repeats (containing CAA or CAT). Two genomic neighbors of HTT [G protein-coupled receptor kinase 4 (GRK4), ∼7 kb upstream; G protein signaling 12 (Rgs12), ∼188 kb downstream] were also unaffected in STHdh cells. (C) mRNA levels of the seven wt human CAG genes and HTT (21 repeats) were also broadly unaffected in HEK293T cells. (D) ZF11xHunt–Kox-1 represses only HTT in a heterozygous human patient-derived cell line (mutant Q45/wt Q21). RNA was extracted from FACS-sorted transduced cells 7 d after infection. (CACNA1A is not expressed in HEK293T cells or the Q45/Q21 patient cell line.)
ZFxHunt Does Not Repress wt CAG-Containing Genes.
Genomes contain several other endogenous CAG-repeat genes; thus, the potential side effects of stably expressed ZFxHunt proteins were assayed by qRT-PCR for atrophin1, ataxin-1, ataxin-2, ataxin-3, ataxin-7, calcium channel α1A subunit, and TATA binding protein (Fig. 4 B–D). No strong effects were seen in either STHdh mouse cells (Fig. 4B) or HEK293T cells (with counterpart human PCR primers) (Fig. 4C). In the latter, even genes with relatively long repeats were unaffected [human HTT, 21 repeats; Tbp, ∼38 repeats (36 CAG repeats broken by two nonconsecutive CAA codons); Table S3].
Because Kox-1 recruits the corepressor KAP-1 and induces long-range repression through the spread of heterochromatin (36), we tested the effects of ZF6xHunt–Kox-1 and ZF11xHunt–Kox-1 on genes neighboring HTT, in stably transduced STHdh cells, by qRT-PCR (Fig. 4B). The two adjacent genes, G protein-coupled receptor kinase 4 (∼7 kb upstream) and G protein signaling 12 (∼188 kb downstream), were both unchanged, suggesting that they are out of range. Thus, ZF6xHunt–Kox-1 and ZF11xHunt–Kox-1 repression appears to be specific for mutant HTT in chromosomal loci. Additionally, a dye-labeling cell viability assay (Fig. S4) revealed no statistically significant toxic effects, encouraging us to test the ZFP’s potential in vivo.
ZF11xHunt–Kox-1 Represses HTT in an HD Patient Cell Line.
Because patients with HD typically have CAG repeats in the range of 42–45 repeats, we tested the potential for repression of these shorter targets in a patient-derived mesothelial cell line (Q45/Q21). We again observed that other genomic CAG-repeat genes remained unaffected, whereas HTT was repressed (Fig. 4D). The Q45/Q21 line is heterozygous and, based on the other repression data, we only expect the Q45 allele to be repressed under these conditions. One hundred percent repression of only the mutant allele would give a maximum twofold repression of HTT in this assay. In fact, we observed an ∼1.8-fold repression of HTT (Fig. 4D). Because the other shorter CAG-repeat genes (including the split 38-repeat Tbp) are not repressed, this indicates that selective inhibition of mutant repeats at the shorter end of the pathological spectrum is possible.
ZFxHunt Fused to Kox-1 Reduces Expression of Mutant HTT in Vivo.
Although ZF11xHunt–Kox-1 apparently functioned well in the stable transfections with dividing cells, these conditions are quite different from the environment of a mammalian brain, where there are many cell types and most of them are nondividing. We therefore decided to test whether any repression could be observed in a more physiological setting.
The rAAV pseudotype 2/1 (AAV2/1), containing AAV2 terminal repeats and AAV1 capsid, efficiently transduces neurons (and most glial and some ependymal cells) and appears to be the most efficient pseudotype for transducing large volumes of the striatum (37). R6/2 mice are one of the most widely used models of HD because they show highly reproducible early-onset symptoms, allowing the use of fewer animals (38). Although R6/2 mice have very long CAG repeats (range: 115–160 repeats), which are rarely found in patients (typically in the range of 42–45 repeats), they are nonetheless ideal to assay acute phenotypic reversal at the molecular, histological, and early-symptom levels (39). Therefore, R6/2 mice were stereotaxically injected at 4 wk of age with AAV2/1-expressing ZF11xHunt–Kox-1.
We initially tested vectors expressing the zinc finger under a CMV promoter, but we observed very low levels of expression of the ZFP mRNA and no repression of mutant HTT. CMV promoters can be quickly silenced by DNA methylation (40), whereas high and persistent levels of expression can be achieved in the CNS with rAAV using the CAG promoter (CMV enhancer, chicken β-actin hybrid promoter) and a Woodchuck hepatitis virus postranscriptional regulatory element (WPRE) (41, 42). We therefore produced a second generation of rAAV using these elements.
CAG-WPRE constructs were injected into the striatum of one brain hemisphere, with AAV2/1-GFP control injections (equal titer) into the other brain hemisphere. ZFP striatal expression was confirmed by Western blot (Fig. S5A), whereas GFP fluorescence showed transduction of large striatal volumes (Fig. 5A). For parallel behavioral studies, both hemispheres were injected (Fig. 5B), also resulting in high and reproducible expression (Fig. 5C). The cellular targeting of AAV2/1 was assayed by immunohistochemistry, showing that transgene expression coincides with a neuronal cell marker, as previously shown by Burger et al. (37) (Fig. S6).
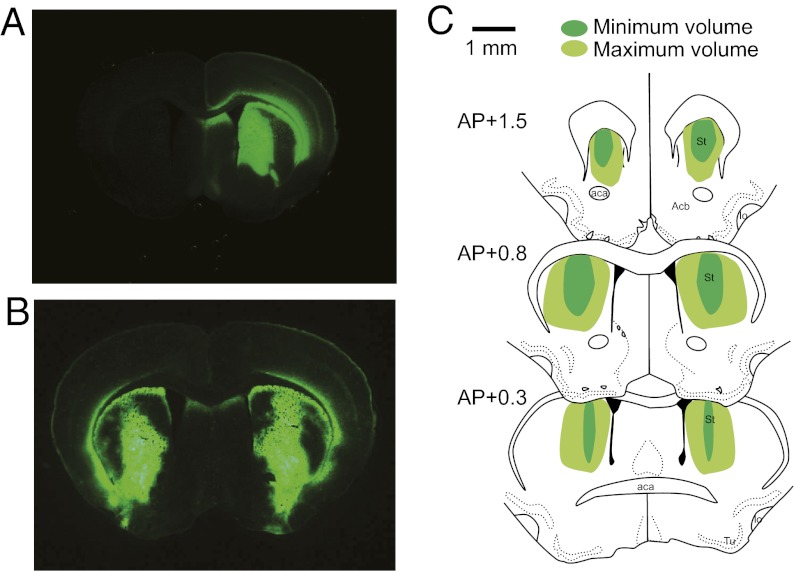
Fig. 5.
Gene delivery by stereotaxis. (A) Cross-section of mouse brain, injected in one hemisphere with AAV2/1-CAG-GFP-WPRE, reveals widespread green fluorescence in the striatum. (B) Similar distributions are seen when injecting the GFP construct in both hemispheres (as for the behavioral assay). The zinc finger construct AAV2/1–CAG–ZF11xHunt–Kox-1–WPRE was injected at an identical titer in one or both hemispheres as described. (C) Schematic drawings show the maximum and minimum volume covered by GFP expression in mice injected in both hemispheres (n = 4). AP levels are as in the Paxinos and Watson atlas of the mouse brain (54). aca, anterior comissure; Acb, nucleus accumbens; AP, anteroposterior; lo, olfactory tract; St, striatum; Tu, olfactory tubercle. (Magnification, A, B: 2.5× objective; Scale bar, C: 1 mm.)
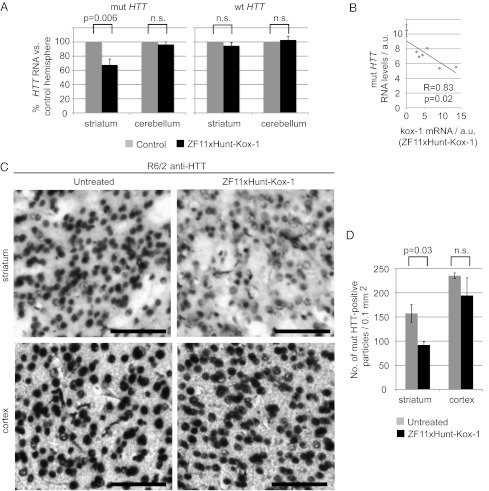
Two weeks after injection, the mice were killed and brain HTT levels were analyzed by qRT-PCR (Fig. 6 A and B). The levels of mutant HTT transgene mRNA were reduced in the striatum by 40% on average (up to 60% in individual mice), compared with the control hemispheres (P = 0.006). Furthermore, both the noninjected cerebellum and wt HTT were unaffected (Fig. 6A). A linear regression analysis comparing expression levels of ZFxHunt–Kox-1 mRNA with those of mutant HTT showed that they correlated closely and negatively (r = 0.83, P = 0.02) (Fig. 6B). Aggregates of mutant HTT protein were detected with anti-HTT antibodies (Fig. 6C), also showing a significant reduction (∼40%) in the treated striatum (Fig. 6D; Student’s t test for paired samples, P = 0.03; Figs. S7 and S8). Again, mutant HTT did not change significantly in the noninjected cortex (Fig. 6 C and D).
Fig. 6.
Zinc finger repression in vivo. (A) qRT-PCR data quantify mRNA levels in mouse samples injected striatally with ZF11xHunt–Kox-1, compared with the control hemisphere. The mutant HTT (mut HTT; n = 6) is repressed ∼40% by the zinc finger construct in the striatum, whereas it is unaffected in the noninjected cerebellum. The wt HTT (n = 9) is unaffected in all samples. Control groups contain both untreated and GFP-treated mice (the groups are similar; Fig. S6B). n.s., not significant. (B) Linear regression analysis of zinc finger expression (ZF11xHunt–Kox-1) vs. mut HTT expression shows a significant negative correlation (P = 0.02). Data are from the treated hemispheres (n = 6) and the corresponding untreated hemispheres (mean ± SEM). a.u., arbitrary units. (C) Anti-HTT immunostaining of brain samples reveals a reduction of mutant aggregates in injected striatum with ZF11xHunt–Kox-1 treatment. (Scale bars: 100 μm.) (D) Quantifying HTT-positive aggregates by automatic counting of mut HTT-positive particles with ImageJ software, as previously described (52) (Fig. S7). The data are from three mice and represent comparisons between their injected and noninjected hemispheres in striatum and cortex.
Neither striatal volume nor cell density in the striatum was affected by treatment or genotype, indicating that the reduction of mutant HTT was not due to a loss of cells and providing no evidence of ZFxHunt toxicity at this level (ANOVA: genotype, P > 0.1; treatment, P > 0.1) (Table S4). Overall, these data are consistent with in vivo dose-dependent repression of mutant HTT by the zinc finger construct.
The results demonstrate acute mutant HTT repression by zinc fingers in the mouse brain. Therefore, we performed in vivo testing to determine if the molecular changes observed in the brain would translate to beneficial effects on the pathological phenotype of the R6/2 HD mouse model.
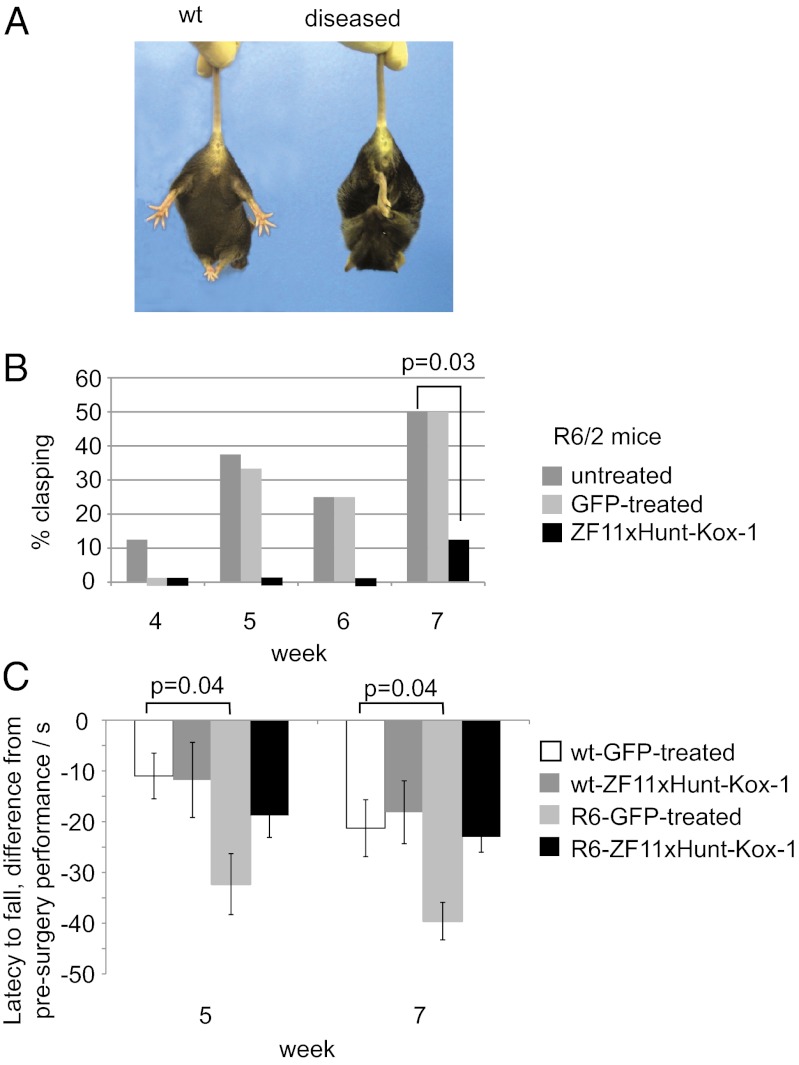
First, we investigated whether the zinc finger repression improved the HD phenotype, using a standard assay in which HD mice display a clasping behavior when lifted by the tail (43) (Fig. 7A). In a double-blind experiment, male R6/2 mice were injected bilaterally in the striata, at 4 wk of age, with AAV2/1–ZF11xHunt–Kox-1 or AAV2/1-GFP. A control group was left unoperated. Strikingly, the mice injected with AAV2/1–ZF11xHunt–Kox-1 showed an almost complete absence of diseased clasping behavior (Fig. 7B). Although control R6/2 mice started displaying clasping as early as week 4, zinc finger-treated R6/2 mice did not clasp until week 7. Even then, only 1 of 8 zinc finger-treated mice clasped, compared with 6 of 12 control R6/2 mice (χ2 test, P = 0.03). GFP-treated R6/2 mice did not differ from unoperated controls (P > 0.7) (Fig. 7B).
Fig. 7.
Rescue of HD phenotypes with in vivo zinc finger treatment. (A) HD mice show a characteristic clasping behavior (diseased) corresponding to neurological pathology (43). (B) Clasping assay shows a significant improvement after zinc finger treatment in both hemispheres (P = 0.03). Only 1 in 8 zinc finger-treated mice displays symptoms by week 7, compared with 6 in 12 control mice. (C) Performance in the accelerating rotarod shows a clear decline with respect to presurgery levels in the GFP-injected R6/2 mice, whereas zinc finger-treated mice do not show a significant decline compared with wt mice.
To assess motor coordination, male R6/2 mice and their wt littermates were tested at 3 wk of age for their baseline performance (latency to fall) in the accelerating rotarod test. They subsequently received bilateral striatal injections at 4 wk, as described above. Repeated-measures ANOVA comparing the decline in R6/2 performance revealed that GFP-treated R6/2 mice were significantly impaired compared with their GFP-treated wt controls (P = 0.035), whereas the R6/2 mice treated with AAV2/1–ZF11xHunt–Kox-1 were not different from the wt mice (P > 0.1) (Fig. 7C).
In our experiments, R6/2 mice do not experience any weight loss or changes in survival compared with their wt counterparts; thus, these phenotypes cannot be used as end points. We did not observe any significant difference in weight between wt and R6/2-treated and untreated mice (Fig. S9), and no mice died during the experiment, either with or without treatment; thus, no conclusion can be drawn about the effect of treatment on these phenotypes.
The data presented establish a proof-of-principle that zinc finger repressors can be active after striatal delivery, with HTT reduction at the RNA and protein level, and significant improvements in disease phenotypes in an HD mouse model.
Discussion
In this study, we have described the design of a ZFP able to recognize and bind both DNA strands of a stretch of CAG repeats, by recognizing both poly-GCA and poly-GCT triplets, to induce transcription repression. As shown in Fig. 2, even naked ZFxHunt is highly efficient episomally and can reduce polyQ-EGFP expression by up to 90%. The mechanism of repression here is likely to be steric hindrance of RNA polymerase complex progression, as reported by Choo et al. (44) for a synthetic ZFP against the bcr-abl oncogene. By contrast, in chromosomal loci, the stronger repression conferred by the Kox-1 repression domain (35, 36) is required to see an effect (Fig. 4).
One major in vitro observation was that longer CAG repeats were repressed more by particular finger constructs under given conditions, suggesting that mass action can be exploited to give a degree of length preference. Binding to different lengths of CAG repeats is, of course, altered by the effective concentrations of both zinc finger and target. We therefore had to find empirically which length of zinc finger and which expression construct would give an effective dose to discriminate between the lengths of CAG repeats in a chosen HD mouse model.
By first using a model cell line derived from striatal cells of a knock-in HD mouse model (the full HTT gene, with variable CAG repeats in exon 1) (34), we found effective constructs and doses that gave a great deal of specificity for repressing mutant huntingtin at a chromosomal level. Although repression of the shorter wt HTT protein is well tolerated for several months in animal models (14, 45), it is desirable to target the mutant allele preferentially (13, 16), and one should not affect other CAG repeat-containing genes. In fact, under the dosages tested, we found conditions in which mutant HTT was strongly repressed for a sustained period of 20 d, with no apparent side effects on other shorter CAG-repeat genes. Therefore, the next step was to explore whether repression could also be achieved in the brain, using gene therapy vectors for delivery.
Gene therapy is an attractive therapeutic strategy for various neurodegenerative diseases. For example, lentiviral vectors have been used to mediate the widespread and long-term expression of transgenes in nondividing cells, such as mature neurons (46). An rAAV vector was also used by Rodriguez-Lebron et al. (47) to deliver anti mutant HTT shRNAs in HD model mice, thereby reducing striatal mutant HTT levels and slowing progression of the HD-like phenotype. As outlined in the introductory section of this report, several strategies to target HTT have shown promise (11–13, 16–19). However, repression in the brain by a synthetic transcription factor has never been reported. It was unknown whether sufficient levels of protein could be produced after in vivo delivery to see any repression, whether transcription repression would be sufficient to reduce protein levels, and whether such constructs would show acute toxicity. In fact, we were able to get effective repressors that were well-tolerated, but this required a degree of further optimization, including the use of a strong CAG promoter and WPRE (42). With these improvements, we observed selective repression of mutant HTT RNA, a reduction in HTT protein aggregates, and an alleviation of associated HD phenotypic deficits.
Overall, the results presented in this study establish a proof-of-principle that zinc fingers can acutely repress mutant HTT in vivo via striatal injection in a dose-dependent manner. These results imply that it is worth pursuing these constructs to test a multitude of possible gene therapy vector and zinc finger variants, with a view toward optimizing longer-term therapeutic benefit.
Materials and Methods
Vectors.
To build a ZFP that recognizes both GCA and GCT DNA sequences, we based our designs on two previous studies. Choo et al. (44) engineered ZFPs with the following α-helical recognition sequences: QAATLQR for GCA and QAQTLQR for GCT. Isalan et al. (48) showed that QRASRKR recognizes GN (T/A). Therefore, we proposed a hybrid design, QRATLQR (ZFxHunt), for GC (T/A) (Fig. 1C). A pUC56 vector containing ZF6xHunt was synthesized by Genscript Corporation. This vector included a T7 promoter; a zif268 DNA-binding domain backbone; an N-terminal nuclear localization signal (PKKKRKV); and restriction sites for deriving ZF4, ZF12, and ZF18 by subcloning (full sequences are given in Dataset S1).
ZF6xHunt was subcloned into the mammalian expression vector pTarget (Promega). A 3 × FLAG tag sequence (DYKDHDG DYKDHDI DYKDDDDK) was introduced by PCR at the N terminus, and either FokI endonuclease domain or Kox-1 (KRAB repression domain) coding sequences were introduced at the C terminus, with a 3 × GGGGS linker.
The plasmid EGFP-HcRed (pEH) vector series was cloned in two steps. First, EGFP coding region was excised from pEGFP-N1 (Clontech), using HindIII/XbaI, and cloned into pGL4.13 (Promega) to give pSV40-EGFP. Then, a PCR product containing CMV-HcRed-polyA and ClaI linkers was cloned into pSV40-EGFP (partially digested with ClaI). The EGFP start codon was mutated to alanine by site-directed mutagenesis, and PCR fragments containing human HTT exon I from different human genomic templates (to obtain different numbers of CAG repeats) were cloned into the pEH EcoRI site, upstream, and in frame with EGFP (pEH-Q series). The pSV40-mCherry vector series were generated by replacing EGFP from the pSV40-EGFP vector series with mCherry using XmaI/XbaI sites.
Gel Shift.
pUC56-ZFxHunt and M13 and M13rev primers were used to generate PCR products for in vitro expression of the indicated ZFP, using the transcription-translation mix (TNT) T7 Quick PCR DNA kit (Promega). dsDNA probes with different numbers of CAG repeats with the standard sequence ACG TAC (CAG)x TCA CAG TCA GTC CAC ACG TC were produced by Klenow fill-in. One hundred nanograms of dsDNA was used in a digoxigenin (DIG)-labeling reaction using a second-generation Gel Shift kit (Roche) following the manufacturer’s instructions. For gel shift assays, 0.005 pmol of DIG-labeled probe was incubated with increasing amounts of TNT-expressed protein in a 20-μL reaction containing 0.1 mg/mL BSA, 0.1 μg/mL poly(deoxyinosinic-deoxycytidylic) acid (poly dI-dC), 5% glycerol, 20 mM Bis-Tris propane, 100 mM NaCl, 5 mM MgCl2, 50 mg/mL ZnCl2, 0.1% Nonidet P-40, and 5 mM DTT, for 1 h at 25 °C. Binding reactions were separated in a 7% (vol/vol) nondenaturing acrylamide gel for 1 h at 100 V and transferred to a nylon membrane for 30 min at 400 mA, and visualization was performed following the manufacturer’s instructions.
Cell Culture and Gene Delivery.
The cell line HEK293T (American Type Culture Collection) was cultured in 5% (vol/vol) CO2 at 37 °C in DMEM (Gibco) supplemented with 10% (vol/vol) FBS (Gibco). Qiagen-purified DNA was transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Briefly, cells were plated onto 10-mm wells to a density of 50% confluency, and 70 ng of reporter plasmid, 330 ng of ZFP expression plasmid, and 2 μL of Lipofectamine 2000 were mixed and added to the cells. Cells were harvested for analysis 48 h later. All assays were performed in triplicate. STHdh+/Hdh+ and STHdhQ111/Hdh111 cells (a gift from M. E. MacDonald, Massachusetts General Hospital, Boston MA) were cultured in 5% (vol/vol) CO2 at 33 °C in DMEM supplemented with 10% (vol/vol) FBS (Gibco) and 400 μg/mL G418 (PAA). For the Q45/Q21 patient-derived cell line, mesothelial cells from a heterozygous (Q45/Q21) patient with HD were collected from urine and cryopreserved with glycerol. After thawing, surviving cells were grown in Chang Medium D (Irvine Scientific). Cells were infected with retroviral particles using the pRetroX system (Clontech) according to the manufacturer’s instructions. Samples were FAC-sorted for GFP expression 7 d after transduction and allowed to recover for 2 wk before extracting RNA for qRT-PCR.
Flow Cytometry Analysis.
Cells were harvested 48 h after transfection and analyzed in a Becton Dickinson FACS Canto Flow cytometer using Becton Dickinson FACSDiva software.
Western Blot.
The 293T cells were harvested 48 h after transfection in 100 μL of 2 × SDS loading dye with Complete protease inhibitor (Roche). Twenty microliters of sample was separated in 4–15% (vol/vol) Criterion Tris⋅HCl ready gels (BioRad) for 2 h at 100 V and transferred to Hybond-C membrane (GE Healthcare) for 1 h at 100 V. Proteins were detected with either the primary antibody anti–β-actin (A1978; Sigma) at a 1:3,000 dilution or anti-EGFP (Roche) at a 1:1,500 dilution and with a peroxidase-conjugated donkey anti-mouse secondary antibody (Jackson ImmunoResearch) at a 1:10,000 dilution. Visualization was performed with an ECL system (GE Healthcare) using a LAS-3000 imaging system (Fujifilm). STHdh cells (from one ∼80% confluent 15-cm plate) were trypsinized and harvested in PBS containing Complete protease inhibitor. Cells were resuspended in radioimmunoprecipitation assay buffer (1% Triton X-100, 1% sodium deoxycholate, 40 mM Tris⋅HCl, 150 mM NaCl, 0.2% SDS, Complete protease inhibitor), incubated on ice for 15 min, and centrifuged at 15,000 × g for 15 min. The supernatant was collected, and the protein concentration was determined using BioRad’s DC protein assay. Sixty micrograms of protein was separated in a 5% (vol/vol) Criterion Tris⋅HCl ready gel (BioRad) for 2 h at 100 V and transferred using an iBlot Dry Blotting System (Invitrogen) for 8 min, and endogenous HTT protein was detected with anti-huntingtin primary antibody (MAB2166; Millipore) at a 1:1,000 dilution.
qRT-PCR.
RNA from cells in one well of an ∼80% confluent 24-well plate was prepared with an RNeasy kit (Qiagen), and 500 ng (unless indicated) was reversed-transcribed with SuperScript II (Invitrogen). A real-time PCR assay was performed using a LightCycler 480 Instrument (Roche) with LightCycler 480 SYBR Green I Master (Roche). SYBR Advantage GC qPCR Premix (Clontech) was used to amplify the human HTT transgene in R6/2 templates. For technical replicates, each PCR assay was performed in triplicate and results were normalized to three housekeeping genes. At least three independent biological replicates (or as indicated if different) were done for each experiment. Primer sets are given in full in Table S3.
Production of AAV.
AAV2/1-CAG-GFP-WPRE and AAV2/1–CAG–ZF11xHunt–Kox-1–WPRE, containing a CAG promoter (CMV early enhancer element and the chicken β-actin promoter), and WPRE were produced at the Centre for Animal Biotechnology and Gene Therapy of the Universitat Autonoma of Barcelona as previously described (49). Recombinant virus was purified by precipitation with PEG 8000, followed by iodixanol gradient ultracentrifugation with a final titer of 7.4 × 1011 genome copies/mL.
Animals.
R6/2 transgenic mice were purchased from the Jackson Laboratories [B6CBA-Tg(HDexon1)62Gpb/3J]. Ovarian-transplanted hemizygous females and wt B6CBAF1/J males were bred in-house, and progeny was genotyped as previously described (50). Animal handling procedures were conducted in accordance with Directive 86/609/EU of the European Commission and following protocols approved by the Ethical Committee of the Barcelona Biomedical Research Park. Stereotaxic injections were performed on 4-wk-old mice. Briefly, mice were anesthetized with a mix of ketamine (75 mg/kg) and medetomidine (1 mg/kg administered i.p.) and fixed on a stereotaxic frame. Analgesia was provided by buprenorphine (8 μg/kg administered s.c.). AAVs were injected bilaterally into the striatum [anterior/posterior, +0.7 mm; medial/lateral, ±1.8 mm; dorsal/ventral (D/V), −3.0 mm relative to bregma] using a 10-μL Hamilton syringe at a rate of 0.25 μL/min controlled by an Ultramicropump (World Precision Instruments). For each hemisphere, a total volume of 3 μL (2.2 × 109 genomic particles) was injected in two steps: 1.5 μL was injected at −3.0 mm D/V, the needle was let to stand for 3 min in position, and the other half was then injected at −2.5 mm D/V. Mice were randomly injected with AAV2/1–CAG–ZF11xHunt–Kox-1–WPRE in one hemisphere and with control AAV expressing GFP in the other hemisphere. Some mice were injected only in one hemisphere with AAV2/1-zinc finger for comparison with the noninjected control hemisphere. Mice were killed after 2 wk for posterior analysis by RT-PCR (primers: mutant HTT, n = 6; wt HTT, n = 9; zinc finger-Kox, n = 9) and histological analyses (wt, n = 3; R6/2, n = 3).
Histology.
Mice were killed with CO2 and rapidly transcardially perfused with saline solution, followed by 75 mL of 4% (wt/vol) paraformaldehyde in 0.01 M PBS. Hemispheres were separated and postfixed in the same fixative overnight at 4 °C and immersed in 30% (wt/vol) sucrose-PBS until they sank. Hemibrains were frozen in a freezing microtome cryostat, and 40-µm-thick coronal sections were obtained. Free-floating sections were collected in six parallel series. One section was stained with H&E for calculation of the volume of the striatum in both R6/2 and wt mice. A second section was processed for immunocytochemical detection of human mutant huntingtin. As controls, we used a mix of slices from wt mice following the whole immunodetection, as well as one slice per hemisphere and R6/2 mouse not incubated with the primary antibody. The rest of the series was frozen in cryoprotectant solution for further use.
Briefly, sections were incubated sequentially in: (i) 1% hydrogen peroxide (H2O2) in Tris-buffered saline (TBS) for 30 min at room temperature for endogenous peroxidase inactivation; (ii) mouse anti-mutant HTT (MAB5374; Millipore) diluted 1:100 in TBS with 0.3% Triton X-100 (Sigma) and 2% (vol/vol) normal goat serum (NGS; Vector Laboratories) overnight at 4 °C; (iii) biotinylated goat anti-mouse IgG (Vector Laboratories) diluted 1:200 in TBS with 0.3% Triton X-100 and 2% (vol/vol) NGS for 2 h at room temperature; and (iv) avidin–biotin–peroxidase complex (ABC Elite Kit; Vector Laboratories) in TBS with 0.3% Triton X-100 for 90 min at room temperature. Following each incubation, sections were washed in TBS (three times for 5 min each time). The resulting peroxidase activity was revealed with FAST diaminobenzidine (Sigma) for 5 min. Sections were rinsed in TBS, mounted onto slides, and cleared with xylene, and coverslips were fixed with Permount (Thermo Fisher Scientific). For NeuN detection, slices were (i) incubated overnight at 4 °C with anti-NeuN (MAB377; Millipore) or (ii) goat–anti-mouse Alexa-555 (A21422; Invitrogen).
Image Analysis.
Determination of the volume and cell density of the striatum.
Striatal volume was determined using Computer Assisted Stereology Toolbox software (Olympus Danmark, A/S) according to the principle of Cavalieri (volume = s1d1 + s2d2 +….sndn) (51) considering eight coronal levels from bregma 1.5-mm level [following the mouse brain atlas (54)] and an interval of 240 μm between the sections. Cell density was calculated in the same slices using the unbiased optical dissector method (53).
Automatic count of mutant HTT-positive particles.
In the mutant HTT immunostained series, four coronal slices per R6/2 mouse and hemisphere from bregma 1.5-mm levels were selected, and a region of interest of 650 × 865 μm2 in the middle of dorsal striatum was captured with a 10× objective using a digital camera attached to an Olympus BX51 microscope. ImageJ software (National Institutes of Health) was used for image analysis. Following the method of Moncho-Bogani et al. (52), counts were obtained and calculated per 0.1 mm2 (Fig. S7). This gave the number of mutant HTT immunoreactive particles in the four slices of both cerebral hemispheres, and the number of particles was averaged, providing a single density measure per mouse.
Clasping Assay.
R6/2 males were bilaterally injected with 3 μL of the same virus in both hemispheres for behavioral assays. Behavioral monitoring was carried out in mice from 4 to 7 wk of age. All the experiments were blindly performed with respect to the treatment of the mice (R6/2 mice: unoperated, n = 8; ZFP-treated, n = 8; and GFP-mock treated, n = 4). Animals were assessed for clasping behavior by suspending them by their tails for 20 s. Mice clasping their hind limbs were given a score of 1, and mice that did not clasp their hind limbs were given a score of 0.
Accelerating Rotarod.
Mice (R6/2 mice: ZFP-treated, n = 12; GFP-mock treated, n = 12; wt mice: ZFP-treated, n = 14; GFP-mock treated, n = 14) were trained at the age of 3 wk to stay on the rod (rotor model LE8200, Panlab, S.L.U., 08940 Barcelona, Spain) at a constant speed of 4 rpm until they reached a criterion of 3 consecutive minutes on it. In the testing phase, mice were put on the rotarod at 4 rpm and the speed was constantly increased for 2 min to 40 rpm. The assay was repeated twice, and the maximum latency to fall from the rod was recorded. Decline in performance was calculated as the difference in the latency to fall from presurgery levels with respect to weeks 5 and 7.
Statistical Analysis.
Error bars in figures are 1 SE based on at least three biological replicates. Data from the repression of polyQ reporter genes experiments were analyzed using a Student’s t test against an expected value of 1 (no repression or activation). Expected percentages of mice clasping were inferred in our population of unoperated R6/2 mice, and a χ2 test for goodness-of-fit was applied to the observed percentages in operated groups. Data from the accelerating rotarod test were analyzed using repeated-measures ANOVA with week as within-subject factor and group as between-subject factor, followed by a post hoc analysis using the Bonferroni correction.
Supplementary Material
Acknowledgments
We thank M. E. MacDonald for the STHdh+/Hdh+ and STHdhQ111/Hdh111 cells. M.I. is funded by the European Research Council (ERC) FP7 201249 ZINC-HUBS and Ministerio de Ciencia e Innovacion (MICINN) BFU2010-17953, and M.D. is funded by Ministerio de Innovación SAF2010-16427 and the Ministerio de Educación y Ciencia-European Molecular Biology Laboratory (MEC-EMBL) agreement.
Footnotes
Conflict of interest statement: M.I. and M.G.-C. are co-authors on a patent associated with this technology.
This article is a PNAS Direct Submission. C.R. is a guest editor invited by the Editorial Board.
See Author Summary on page 18264 (volume 109, number 45).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206506109/-/DCSupplemental.
References
- 1.Walker FO. Huntington’s Disease. Semin Neurol. 2007;27(2):143–150. doi: 10.1055/s-2007-971176. [DOI] [PubMed] [Google Scholar]
- 2.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 3.Cha JH. Transcriptional signatures in Huntington’s disease. Prog Neurobiol. 2007;83(4):228–248. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar P, Kalonia H, Kumar A. Huntington’s disease: Pathogenesis to animal models. Pharmacol Rep. 2010;62(1):1–14. doi: 10.1016/s1734-1140(10)70238-3. [DOI] [PubMed] [Google Scholar]
- 5.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 6.Ramaswamy S, Kordower JH. Gene therapy for Huntington’s disease. Neurobiol Dis. 2012;48(2):243–254. doi: 10.1016/j.nbd.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Sharp AH, et al. Widespread expression of Huntington’s disease gene (IT15) protein product. Neuron. 1995;14(5):1065–1074. doi: 10.1016/0896-6273(95)90345-3. [DOI] [PubMed] [Google Scholar]
- 8.Duyao MP, et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995;269(5222):407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 9.Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26(3):300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 10.Matsui M, Corey DR. Allele-selective inhibition of trinucleotide repeat genes. Drug Discov Today. 2012;17(9-10):443–450. doi: 10.1016/j.drudis.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Bilsen PH, et al. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington’s disease patient-derived fibroblasts. Hum Gene Ther. 2008;19(7):710–719. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Engelman J, Friedlander RM. Allele-specific silencing of mutant Huntington’s disease gene. J Neurochem. 2009;108(1):82–90. doi: 10.1111/j.1471-4159.2008.05734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfister EL, et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Curr Biol. 2009;19(9):774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grondin R, et al. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain. 2012;135(Pt 4):1197–1209. doi: 10.1093/brain/awr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McBride JL, et al. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol Ther. 2011;19(12):2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, et al. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat Biotechnol. 2009;27(5):478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, Matsui M, Corey DR. Allele-selective inhibition of mutant huntingtin by peptide nucleic acid-peptide conjugates, locked nucleic acid, and small interfering RNA. Ann N Y Acad Sci. 2009;1175:24–31. doi: 10.1111/j.1749-6632.2009.04975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kordasiewicz HB, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74(6):1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer PO, et al. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol. 2010;28(3):256–263. doi: 10.1038/nbt.1608. [DOI] [PubMed] [Google Scholar]
- 20.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 21.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 22.Jantz D, Amann BT, Gatto GJ, Jr, Berg JM. The design of functional DNA-binding proteins based on zinc finger domains. Chem Rev. 2004;104(2):789–799. doi: 10.1021/cr020603o. [DOI] [PubMed] [Google Scholar]
- 23.Sera T, Uranga C. Rational design of artificial zinc-finger proteins using a nondegenerate recognition code table. Biochemistry. 2002;41(22):7074–7081. doi: 10.1021/bi020095c. [DOI] [PubMed] [Google Scholar]
- 24.Mandell JG, Barbas CF., 3rd Zinc Finger Tools: Custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34(Web Server issue):W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JS, Lee HJ, Carroll D. Genome editing with modularly assembled zinc-finger nucleases. Nat Methods. 2010;7(2) doi: 10.1038/nmeth0210-91b. 91, author reply 91–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19(7):1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurt JA, Thibodeau SA, Hirsh AS, Pabo CO, Joung JK. Highly specific zinc finger proteins obtained by directed domain shuffling and cell-based selection. Proc Natl Acad Sci USA. 2003;100(21):12271–12276. doi: 10.1073/pnas.2135381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright DA, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1(3):1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 29.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31(2):294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittelman D, et al. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc Natl Acad Sci USA. 2009;106(24):9607–9612. doi: 10.1073/pnas.0902420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore M, Klug A, Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc Natl Acad Sci USA. 2001;98(4):1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JS, Pabo CO. Getting a handhold on DNA: Design of poly-zinc finger proteins with femtomolar dissociation constants. Proc Natl Acad Sci USA. 1998;95(6):2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trettel F, et al. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9(19):2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- 35.Margolin JF, et al. Krüppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci USA. 1994;91(10):4509–4513. doi: 10.1073/pnas.91.10.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groner AC, et al. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6(3):e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burger C, et al. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10(2):302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 38.Crook ZR, Housman DE. Dysregulation of dopamine receptor D2 as a sensitive measure for Huntington disease pathology in model mice. Proc Natl Acad Sci USA. 2012;109(19):7487–7492. doi: 10.1073/pnas.1204542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gil JM, Rego AC. The R6 lines of transgenic mice: A model for screening new therapies for Huntington’s disease. Brain Res Brain Res Rev. 2009;59(2):410–431. doi: 10.1016/j.brainresrev.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Migliaccio AR, et al. Stable and unstable transgene integration sites in the human genome: Extinction of the Green Fluorescent Protein transgene in K562 cells. Gene. 2000;256(1-2):197–214. doi: 10.1016/s0378-1119(00)00353-x. [DOI] [PubMed] [Google Scholar]
- 41.Garg S, Oran AE, Hon H, Jacob J. The hybrid cytomegalovirus enhancer/chicken beta-actin promoter along with woodchuck hepatitis virus posttranscriptional regulatory element enhances the protective efficacy of DNA vaccines. J Immunol. 2004;173(1):550–558. doi: 10.4049/jimmunol.173.1.550. [DOI] [PubMed] [Google Scholar]
- 42.Tenenbaum L, et al. Recombinant AAV-mediated gene delivery to the central nervous system. J Gene Med. 2004;6(Suppl 1):S212–S222. doi: 10.1002/jgm.506. [DOI] [PubMed] [Google Scholar]
- 43.Reddy PH, et al. Transgenic mice expressing mutated full-length HD cDNA: A paradigm for locomotor changes and selective neuronal loss in Huntington’s disease. Philos Trans R Soc Lond B Biol Sci. 1999;354(1386):1035–1045. doi: 10.1098/rstb.1999.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choo Y, Sánchez-García I, Klug A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature. 1994;372(6507):642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 45.Boudreau RL, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol Ther. 2009;17(6):1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreyer JL. Lentiviral vector-mediated gene transfer and RNA silencing technology in neuronal dysfunctions. Methods Mol Biol. 2010;614:3–35. doi: 10.1007/978-1-60761-533-0_1. [DOI] [PubMed] [Google Scholar]
- 47.Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington’s disease transgenic mice. Mol Ther. 2005;12(4):618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isalan M, Klug A, Choo Y. Comprehensive DNA recognition through concerted interactions from adjacent zinc fingers. Biochemistry. 1998;37(35):12026–12033. doi: 10.1021/bi981358z. [DOI] [PubMed] [Google Scholar]
- 49.Salvetti A, et al. Factors influencing recombinant adeno-associated virus production. Hum Gene Ther. 1998;9(5):695–706. doi: 10.1089/hum.1998.9.5-695. [DOI] [PubMed] [Google Scholar]
- 50.Benn CL, et al. Genetic knock-down of HDAC7 does not ameliorate disease pathogenesis in the R6/2 mouse model of Huntington’s disease. PLoS ONE. 2009;4(6):e5747. doi: 10.1371/journal.pone.0005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai Y, Dudek NL, Li Q, Fowler SC, Muma NA. Striatal expression of a calmodulin fragment improved motor function, weight loss, and neuropathology in the R6/2 mouse model of Huntington’s disease. J Neurosci. 2009;29(37):11550–11559. doi: 10.1523/JNEUROSCI.3307-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moncho-Bogani J, Martinez-Garcia F, Novejarque A, Lanuza E. Attraction to sexual pheromones and associated odorants in female mice involves activation of the reward system and basolateral amygdala. Eur J Neurosci. 2005;21(8):2186–2198. doi: 10.1111/j.1460-9568.2005.04036.x. [DOI] [PubMed] [Google Scholar]
- 53.Oorschot DE. Total number of neurons in the neostriatal, pallidal, subthalamic, and substantia nigral nuclei of the rat basal ganglia: A stereological study using the cavalieri and optical disector methods. J Comp Neurol. 1996;366(4):580–599. doi: 10.1002/(SICI)1096-9861(19960318)366:4<580::AID-CNE3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 54.Paxinos G, Franklin . 2001. The mouse brain in stereotaxic coordinates. 2nd Edition (Academic Press) [Google Scholar]