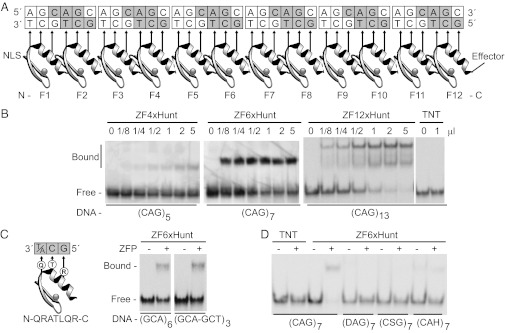
Fig. 1.
Zinc finger arrays to bind CAG repeats. (A) A twelve-finger array shows recognition helices contacting 5′-GCT-3′ bases on the lower DNA strand. Similar arrays of 4, 6, 12, or 18 zinc fingers were built (ZF4xHunt, ZF6xHunt, ZF12xHunt, and ZF18xHunt). Nuclear localization signals (NLS) and effectors (e.g., Kox-1 transcription repression domain) were added to N and C termini, respectively. (B) Gel shift assays show 4-, 6-, or 12-finger arrays binding poly-CAG DNA and forming distinct complexes. TNT is a negative control. (C) A hybrid zinc finger design recognizes 5′-GC(A/T)-3′, allowing binding to either the (GCA)n or (GCT)n complementary strand of the CAG repeat. A gel shift assay shows equal binding to GCA or GCT triplets in mixed sequences. (D) Specificity gel shift assay. Zinc fingers bind preferentially to CAG repeats (CAG7) compared with degenerate mutant sequences (DAG7, CSG7, or CAH7; D = A, G, T; S = C, G; H = A, C, T).