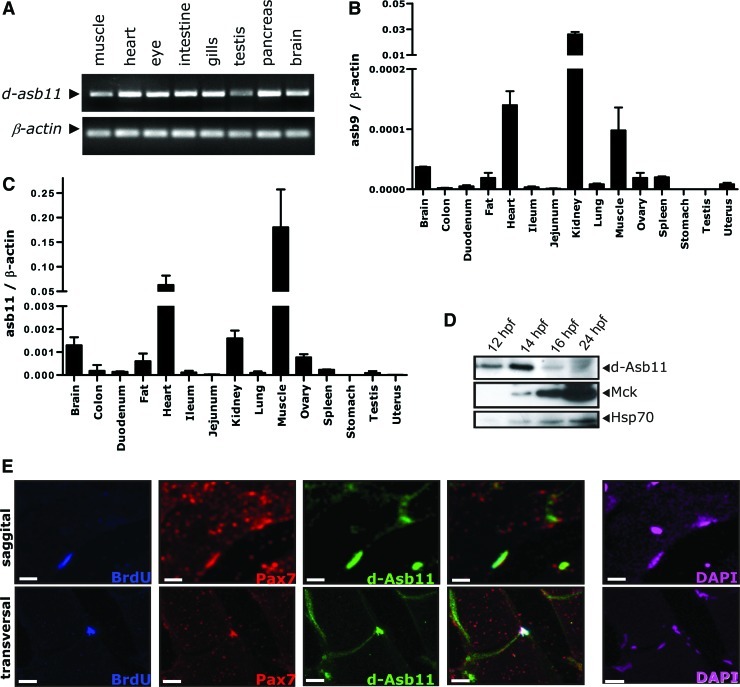
FIG. 1.
Distribution of Asb11 expression in different tissues and colocalization with Pax7+ satellite cells. (A) RT–polymerase chain reaction analysis of zebrafish d-Asb11 (asb11) in mRNA samples isolated from muscle, heart, eye, intestine, gill, testis, pancreas, and brain of adult fish. Zebrafish β-actin was used as template control. (B, C) Various types of tissue from adult mice were analyzed for mRNA expression of murine Asb9 (Asb9) (B) and Asb11 (Asb11) (C). Results were normalized against β-actin expression. Asb9 is showing the highest expression in kidney, heart, and muscle. Asb11 shows the highest expression in muscle, heart, kidney, and brain. The bars indicate the standard error of the mean and were calculated from the results of 3 individual experiments. (D) Analysis of protein levels of Asb11 and muscle creatine kinase (MCK), a marker of muscle terminal differentiation, in different stages during embryogenesis. Hsp70 expression is used as input control. Expression of MCK and Asb11 showed inverse correlation during zebrafish embryogenesis. The images shown are representative for 3 independent experiments. (E) Adult zebrafish muscle was immunostained with bromodeoxyuridine (BrdU), Pax7, and d-Asb11 antibodies. Top panels show a representative staining of sagittal sections and bottom panels show a representative of transverse sections. d-Asb11 revealed a distinct expression pattern on the sarcolemma where it colocalizes with Pax7+ cells. The capacity of Pax7+/Asb11+ cells for long-term BrdU incorporation was tested and confirmed that this double-positive compartment truly represents slow-cycling satellite cells. 4′,6-diamidino-2-phenylindole staining was included to determine cell locations (white bars are 10 μm). Color images available online at www.liebertpub.com/scd