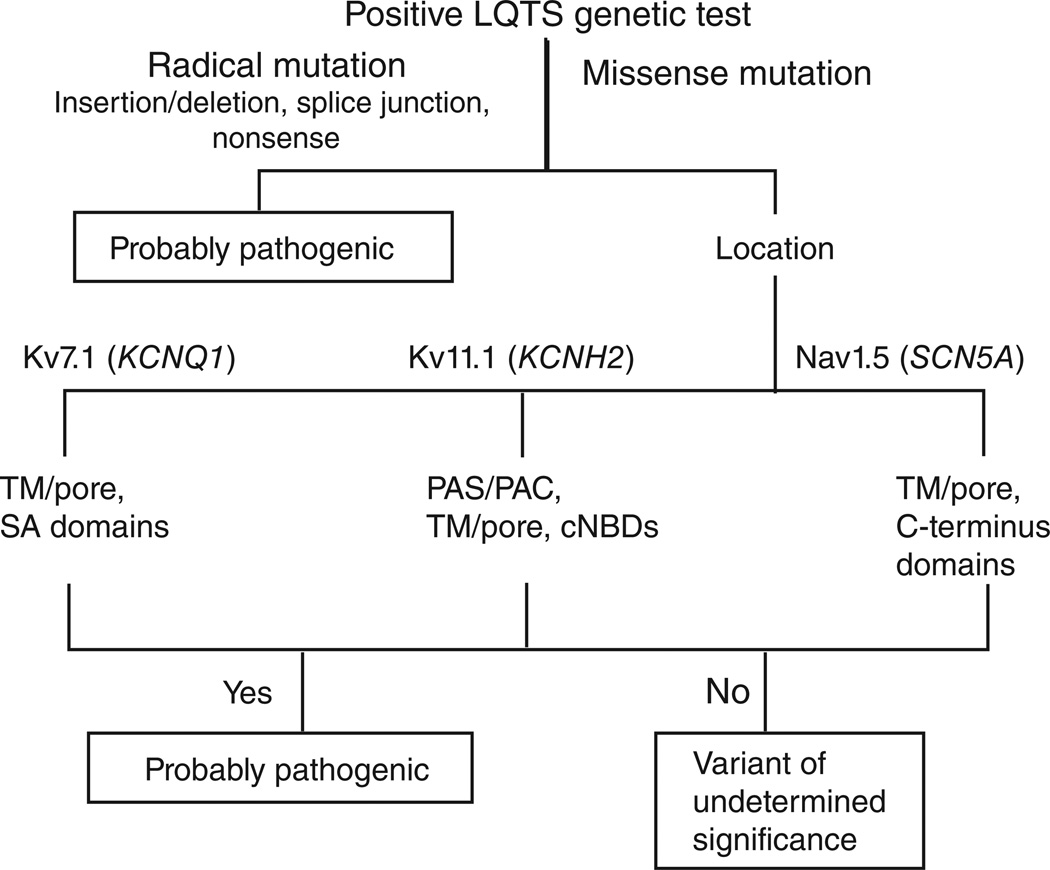
Figure 1.
Algorithm for interpreting a positive genetic test for long QT syndrome (LQTS). Radical mutations that significantly alter the coding protein, such as insertions or deletions, alteration of the intron or exon splice sites, and nonsense mutations that introduce a premature protein truncation, are probably LQTS-associated. Missense mutations localizing to KCNQ1-encoded Kv7.1 (within the TM/ linker/pore or the SA domain), KCNH2-encoded Kv11.1 (within the PAS/PAC, TM/linker/pore, or the cNBD domains), and SCN5A-encoded Nav1.5 (within the TM/linker or the C-terminal domains) are associated with LQTS. Mutations localizing outside these specific domains of Kv11.1 and Nav1.5 are of uncertain disease significance. cNBD, cyclic nucleotide-binding domain; PAS/PAC, per-arnt-sim–associated C-terminal; SA, subunit assembly; TM, transmembrane.