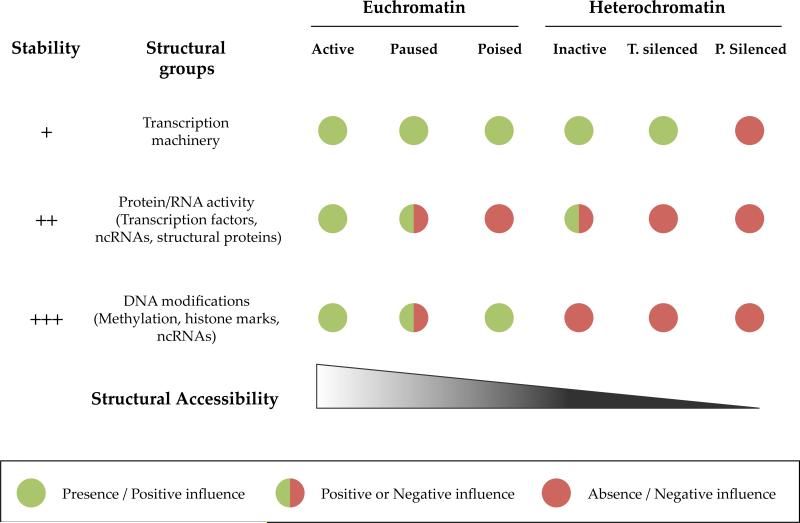
Figure 2. Hypothesized structural differences between chromatin states.
We hypothesize that three key structural groups can, in conjunction with endogenous 3D genomic structure, distinguish the different proposed states at the level of the gene, namely the accessibility of the gene to RNA polymerase II and transcriptional machinery binding (top row), the presence of the appropriate transcription factor in the nucleus and the localization of specific chromatin structural proteins/non- coding RNAs to the gene (middle row) and the presence of conducive, and absence of inhibitory, DNA and histone modifications at the gene (bottom row). Together these help define the structural accessibility of the gene (bottom triangle, with decreasing accessibility as one moves from left to right through the states in as displayed). For each state we propose which class of structural elements are conducive to transcription (green) and which are limiting (red). We further propose that for two states in particular, paused and inactive, more experimental data is needed to determine which class of structural features cause the different transcriptional read-outs (half red/half green demarks possibilities which must be confirmed experimentally). Finally, for each class of structural groups we propose their relative stability and thus the energy required to interconvert between states when these levels of structural regulation are modified.