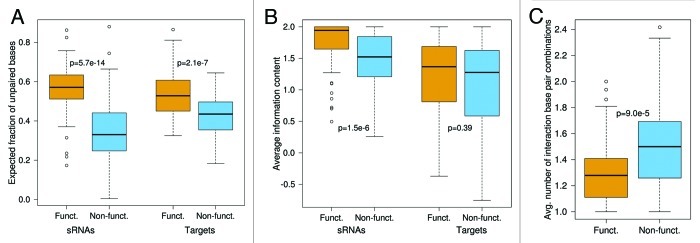
Figure 2. Comparison of interaction site features between functional (orange) and non-functional (blue) interactions. The plots show the (A) interaction site accessibility, (B) interaction site sequence conservation and (C) average number of different interaction base pairings, respectively. Interaction sites both in sRNAs and targets are significantly more accessible in functional than in non-functional interactions. In contrast, only sRNA interaction sites show significant evolutionary conservation. The average number of intermolecular base pair combinations is significantly smaller in the functional interactions. All p-values were calculated by Wilcoxon rank sum test.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.