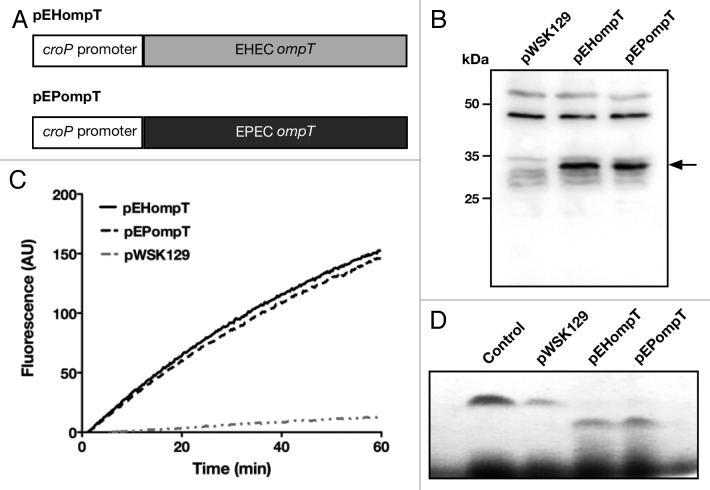
Figure 1. EHEC and EPEC OmpT degrade peptides at similar rates (A) Schematic of EHEC and EPEC ompT expressed from the croP promoter. Plasmids pEHompT and pEPompT are derived from pWSK129 and contain the C. rodentium croP promoter fused to the ompT open-reading frames of EHEC or EPEC, respectively. (B) Amounts of EHEC and EPEC OmpT produced by C. rodentium ΔcroP cells transformed with pEHompT or pEPompT were analyzed by western blotting using an antiserum developed against the CroP OM protease of C. rodentium (74% identical to OmpT).14 (C) Cleavage of the synthetic FRET substrate containing the dibasic sequence RK in its center [2Abz-SLGRKIQI-K(Dnp)-NH2] was monitored over time by measuring fluorescence emission at 430 nm. (D) Proteolytic degradation of LL-37 by C. rodentium ΔcroP cells expressing EHEC or EPEC ompT. LL-37 was incubated for 1 h with the various bacterial strains. Aliquots were separated by Tris-Tricine SDS-PAGE (10–20% acrylamide) and gels were stained with Coomassie blue G-250. The control lane, in which no bacteria were added, contained 1.5 µg LL-37 in phosphate-buffered saline.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.