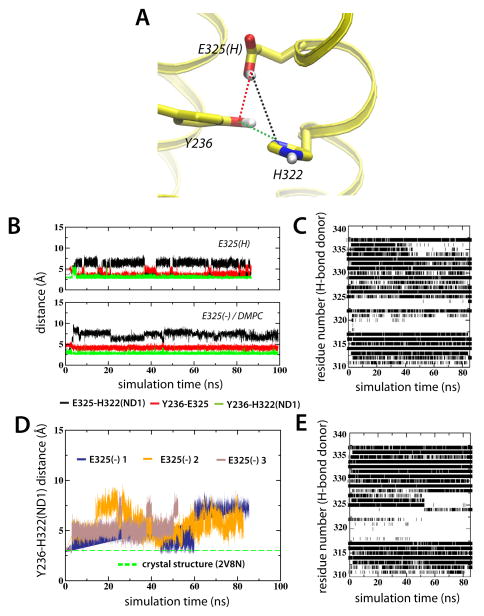
Figure 6.
Deprotonation of Glu325 affects the solvation shell surrounding His322. (A) Glu325, His322, Tyr236 interactions displayed using the average configuration of the last ns of the E325(H) simulation. (B) The dynamics of Glu325, Tyr236 and His322 are represented as interatomic distances between Glu325-His322 (black), Glu325-Tyr236 (red) and His322-Tyr236 (green). (C) Backbone hydrogen bonding of helix X for the E325(H) simulation. O-HN hydrogen bonds between residue i and i+4 were defined as existing when the O-N distance was within 3.5 Å and the O-H-N angle was greater than 130°. The ev olution of the Y236-H322(ND1) interatomic distance for E325(−) 1, E325(−) 2 and E325 (−) 3 in POPE are shown in (D). (E) corresponds to (C) for the E325(−) 1 simulation.