Figure 1.
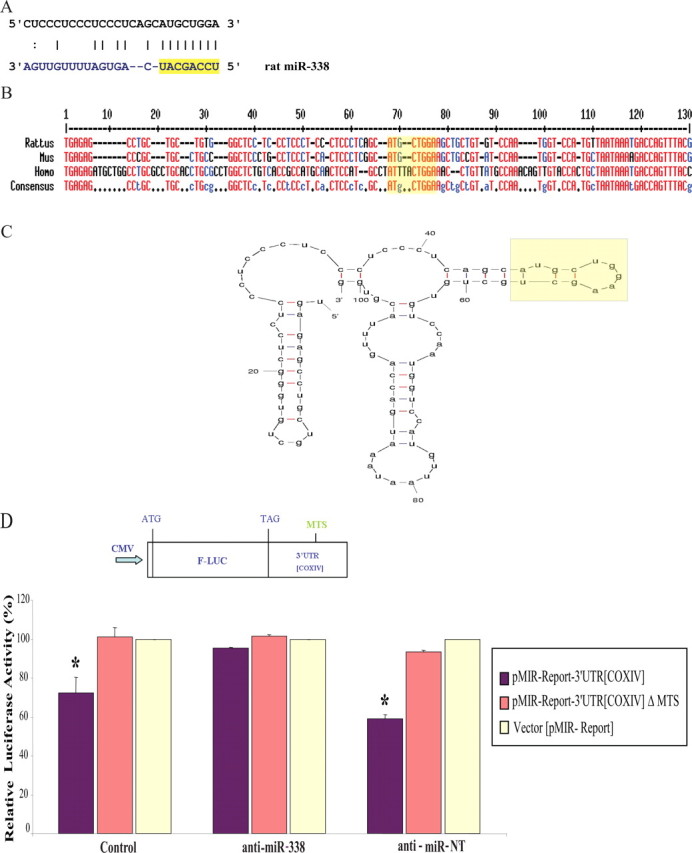
MiR-338 targets rat COXIV. A, Putative miR-338 binding site in the rat COXIV 3′UTR. The 8 nt seed sequence is marked in yellow. B, C, Sequence (B) and secondary structure (C) of the COXIV 3′UTR, as determined by secondary structure prediction analysis (Mfold). The miR-338 target site (MTS) is indicated in yellow. D, The luciferase reporter plasmids carrying the firefly luciferase coding sequence (F-LUC) attached to the entire 3′UTR of rat COXIV (wild-type) or the 3′UTR in which the miR-338 target site was deleted (ΔMTS) are diagrammed. Luciferase expression was driven by the cytomegalovirus promoter (CMV). Luciferase activity of wild-type or ΔMTS COXIV 3′UTR reporter genes in the absence (control) or presence of the anti-miR-338 (25 nm), or a nontargeting (NT) anti-miR oligonucleotide (25 nm), is shown. Luciferase reporter constructs were cotransfected in equal ratios with the control plasmids coding for β-galactosidase (0.5 μg for each plasmid per transfection). The ratio of reporter to control plasmids in relative luminescence units was normalized for each reporter and plotted as a percentage of sham-transfected control value. Anti-miR oligonucleotide transfections were performed within 4 h following plasmid DNA transfection. Error bars represent the SEM for n = 10 cultures. *p < 0.05.
