Table 1.
Comparison of different approaches on the four SNPs identified in Nejentsev et al.’s article of which the first two SNPs within IFIH1 gene were replicated in their follow-up genotyping experiment
| SNP | Location | Chr. | Alleles |
P-value |
|||
|---|---|---|---|---|---|---|---|
| Nejentsev’s Fisher’s | Our Fisher’s | Our EB1 | Our EB2 | ||||
| rs35337543 | IFIH1, intron 8 | chr2 | G C C |
0.000044 | 0.0032 | 0.0029 | 0.016 |
| rs35667974 | IFIH1, exon 14 | chr2 | A G G |
0.0049 | 0.076 | 0.017 | 0.011 |
| ss107794688 | CLEC16A, intron 23 | chr16 | C T T |
0.016 | 0.045 | 0.0047 | 0.034 |
| ss107794687 | CLEC16A, intron 11 | chr16 | C T T |
0.023 | 0.000046 | 0.00021 | 0.000005 |
Since different alignment software and filters were used, our Fisher’s exact results were slightly different from Nejentsev et al.rsquo;s. Overall, all the four SNPs still showed significance in our preferred EB2 model (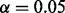 ).
).
