Table 2.
The SNPs identified by EB2 model after initial filtering
| SNP | Gene | Alleles | MAF |
P-value |
OR | T1D SNP | r2 | ||
|---|---|---|---|---|---|---|---|---|---|
| Control | Case | Fisher’s | EB2 | ||||||
| rs3184504 | SH2B3 | T C C |
0.53 | 0.41 | 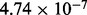 |
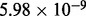 |
0.62 | rs3184504 | 1 |
| rs72650660 | CLEC16A | C T T |
0.043 | 0.011 | 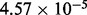 |
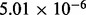 |
0.25 | – | – |
| rs8052325 | CLEC16A | A G G |
0.13 | 0.082 | 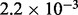 |
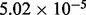 |
0.60 | rs12708716 | 0.26 |
| rs2476601 | PTPN22 | G A A |
0.093 | 0.15 | 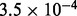 |
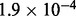 |
1.72 | rs2476601 | 1 |
| rs3827734 | PTPN22 | A T T |
0.0025 | 0.020 | 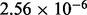 |
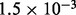 |
8.14 | – | – |
| rs1800521 | AIRE | T C C |
0.23 | 0.32 | 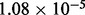 |
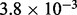 |
1.58 | rs760426 | 0.04 |
| rs11594656 | 0.0029 | ||||||||
| rs28360489 | IL2RA | C T T |
0.11 | 0.084 | 0.039 | 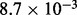 |
0.74 | rs2104286 | 0.10 |
| rs12722495 | 0.62 | ||||||||
OR is odds ratio of minor allele for T1D. r2 is the correlation coefficient with the most associated independent T1D SNPs (marker SNPs that have been identified associated with T1D before) in each region (http://www.t1dbase.org/page/Regions).
