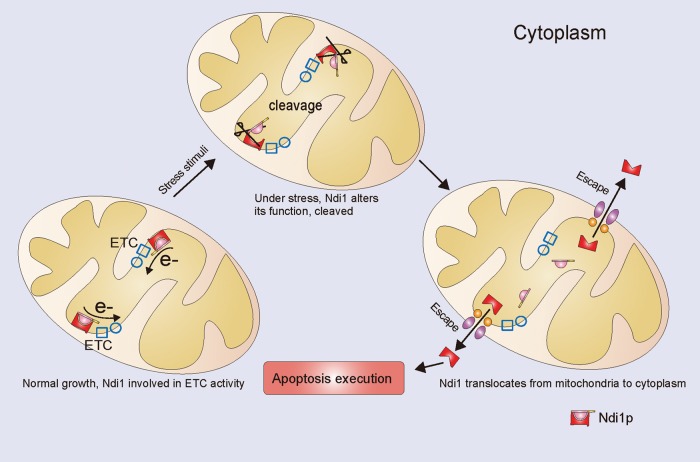
Ndi1, the yeast homologue of caspase-independent apoptosis inducer AMID, turns out to be a general, as well as a potent, yeast apoptotic factor. This protein normally acts at the first step in respiration but, when stressed, cleaves its protective N-terminal, escapes from the mitochondria, and switches to become apoptotic.
Abstract
Saccharomyces cerevisiae NDI1 codes for the internal mitochondrial ubiquinone oxidoreductase, which transfers electrons from NADH to ubiquinone in the respiratory chain. Previously we found that Ndi1 is a yeast homologue of the protein apoptosis-inducing factor–homologous mitochondrion-associated inducer of death and displays potent proapoptotic activity. Here we show that S. cerevisiae NDI1 is involved in apoptosis induced by various stimuli tested, including H2O2, Mn, and acetate acid, independent of Z-VAD-fmk (a caspase inhibitor) inhibition. Although Ndi1 also participates in respiration, its proapoptotic property is separable from the ubiquinone oxidoreductase activity. During apoptosis, the N-terminal of Ndi1 is cleaved off in the mitochondria, and this activated form then escapes out to execute its apoptotic function. The N-terminal cleavage appears to be essential for the manifestation of the full apoptotic activity, as the uncleaved form of Ndi1 exhibits much less growth-inhibitory activity. Our results thus indicate an important role of Ndi1 in the switch of life and death fates in yeast: during normal growth, Ndi1 assimilates electrons to the electron transport chain and initiates the respiration process to make ATP, whereas under stresses, it cleaves the toxicity-sequestering N-terminal cap, is released from the mitochondria, and becomes a cell killer.
INTRODUCTION
Apoptosis is an essential bioprocess for removing unwanted or damaged cells. It is a genetically controlled and highly conserved process shared not only by metazoan organisms, but also by unicellular organisms such as the budding yeast Saccharomyces cerevisiae. As a unicellular organism and an excellent model of genetics, yeast is useful for apoptosis studies (Greenwood and Ludovico, 2010). Although much is known about the apoptosis program in multicellular organisms, relatively less is characterized in budding yeast. Nevertheless, apoptosis in yeast has been found to resemble that in metazoans in both morphological and molecular aspects (Madeo et al., 1997, 1999; Carmona-Gutierrez et al., 2010). In particular, when yeast is under stress, typical markers of apoptosis such as DNA fragmentation, phosphatidylserine externalization, and chromatin condensation are observed. Apoptotic yeast also exhibit prominent Z-VAD-fmk–binding activity. In addition, yeast has a caspase homologue, Yca1. Although whether Yca1 possesses canonical caspase activities as in metazoans is in dispute, the fact that it is an important player in yeast apoptosis is unquestioned (Madeo et al., 2002; Watanabe and Lam, 2005).
Mitochondria are crucial organelles for both energy production and apoptosis mediation (Green and Reed, 1998; Kroemer et al., 2007; Abdelwahid et al., 2011). In metazoans, two mitochondrial proteins, apoptosis-inducing factor (AIF) and AIF-homologous mitochondrion-associated inducer of death (AMID), are involved in the caspase-independent apoptotic pathway (Susin et al., 1999; Wu et al., 2002). Just like several other key players of apoptosis, AIF and AMID homologues have also been found in budding yeast (Wissing et al., 2004; Li et al., 2006). Yeast Ndi1, a ubiquinone oxidoreductase, which assimilates electron from NADH to ubiquinone in the respiratory chain, displays the most sequence homology (28% identity overall) to AMID and slightly less to AIF. It is known that under stress AIF protein is cleaved and moves out of the mitochondrion to the nucleus to initiate apoptosis (Otera et al., 2005). Compared to AIF, AIMD is a less well characterized protein. In previous work, we found that NDI1 also displays apoptotic activity (Li et al., 2006). Here we further characterize the molecular mechanism of Ndi1 in promoting apoptosis.
RESULTS
Ndi1 mediates apoptosis induced by various stresses and is functionally independent of yeast Z-VAD-fmk–binding activity
We previously showed that Ndi1 overexpression induces yeast apoptosis and is involved in aging-related apoptosis. To determine whether NDI1 is involved in apoptosis induced by other external stress factors, NDI1-overexpressing or deletion strains were subjected to H2O2 (Madeo et al., 1999) and Mn (Liang and Zhou, 2007). Low doses of H2O2 and Mn are excellent yeast apoptosis inducers. The viability test results showed that the strain overexpressing NDI1 survived worse than the corresponding vector control (Figure 1, A and D). In contrast, the deletion mutant of NDI1 was resistant to Mn- and H2O2-induced apoptosis compared with wild-type yeast (Figure 1, B and C). In addition, we found that Ndi1 is involved in apoptosis induced by acetic acid (Ludovico et al., 2001), another commonly used apoptosis inducer (unpublished data).
FIGURE 1:
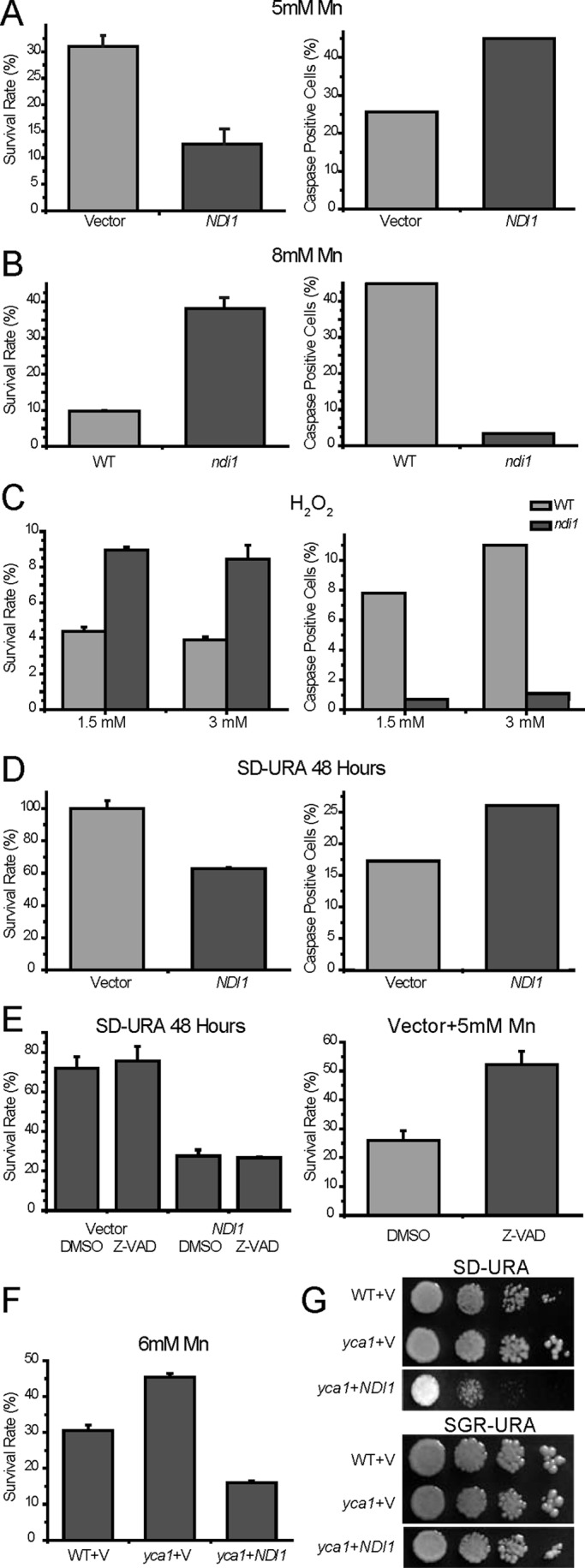
NDI1 participates in apoptosis induced by various stimuli. (A, B) NDI1 is involved in Mn-induced apoptosis. NDI1 overexpression and deletion resulted respectively in reduced and increased viabilities under Mn stress, accompanied by changes of caspase-like activity. Yeast cells were grown on SGR-URA medium to a low cell density and treated for 12 h with 5 or 8 mM Mn. For each culture, a fraction was quantified for survival; another fraction was labeled for active caspase with FITC-VAD-fmk and analyzed by flow cytometry. (C) NDI1 is also involved in H2O2-induced apoptosis. Survival rates and active caspase signals of wild-type and ndi1 mutant treated with classic apoptosis inducer H2O2 at concentrations of 1.5 and 3 mM for 100 min. (D) NDI1 overexpression by itself results in apoptosis when grown in respiration-restricted media (glucose), also associated with an increase of caspase-like activity. (E) The increased caspase-like activity plays an insignificant role in the apoptosis resulting from Ndi1 overexpression. Left, survival rates of Ndi1 and vector-transformed yeast cells with or without preincubation of 400 μM Z-VAD-fmk for 30 min (before being cultured in SD-URA for 48 h). Dimethyl sulfoxide (DMSO) serves as the blank control. Right, the positive control to indicate that Z-VAD (100 μM) is functional, as it can increase cell viability when insulted by Mn. (F, G) NDI1-mediated apoptosis is independent of YCA1. NDI1 overexpression in yca1-deletion mutant similarly aggravated apoptosis, as shown by survival test and spotting assay. Survival data are presented as the mean ±SEM of three independent experiments.
To determine whether the yeast cells undergoing NDI1-mediated apoptosis display increased endogenous metacaspase activity, we assayed the VAD-binding activity (an assay for potential metacaspase activity) in the NDI1-overexpressing and deletion mutant strains. After treatment by various stresses, yeast cells were incubated with a fluorescein isothiocyanate (FITC)–conjugated caspase inhibitor (FITC-VAD-fmk) probe, which can bind and abrogate activated metazoan caspases irreversibly, and then analyzed by flow cytometry. Of interest, strains overexpressing NDI1 accumulated more FITC-VAD-fmk (Figure 1, A and D), whereas ndi1 mutants were labeled less (Figure 1, B and C).
As a homologue of AMID, which is known to participate in apoptosis in a caspase-independent manner, it is very important to establish whether the Ndi1-mediated apoptosis is caspase dependent, in other words, whether the increased VAD-binding signal associated with Ndi1-induced apoptosis is functional. To address this question, we analyzed the survival rates of the NDI1-overexpressing strain with or without preincubation with the pan-caspase inhibitor Z-VAD-fmk. The addition of Z-VAD-fmk did not suppress the apoptotic phenotype due to Ndi1 overexpression (Figure 1E). As a control to show that Z-VAD-fmk is working, we stressed wild-type yeast with Mn with or without Z-VAD-fmk. Mn-induced apoptosis was previously shown to be partially dependent on Yca1 and associated with increased VAD-binding signal (Liang and Zhou, 2007). Very clearly, Z-VAD-fmk could significantly abrogate cell death caused by Mn (Figure 1E).
Because Yca1 is the only known yeast metacaspase, to demonstrate genetically that Ndi1 functions independently of Yca1, we overexpressed NDI1 in yca1 mutant background. Consistently, NDI1 overexpression exacerbated cell death in the absence of Yca1 (Figure 1, F and G).
The results show that Ndi1 plays an important role in Mn- and H2O2-induced apoptosis. Although accompanied by increase in VAD-binding signal, Ndi1-induced apoptosis appears to be functionally independent of the potential yeast metacaspase activity.
Proapoptotic activity of Ndi1 is separable from its ubiquinone oxidoreductase activity and independent of electron transport chain
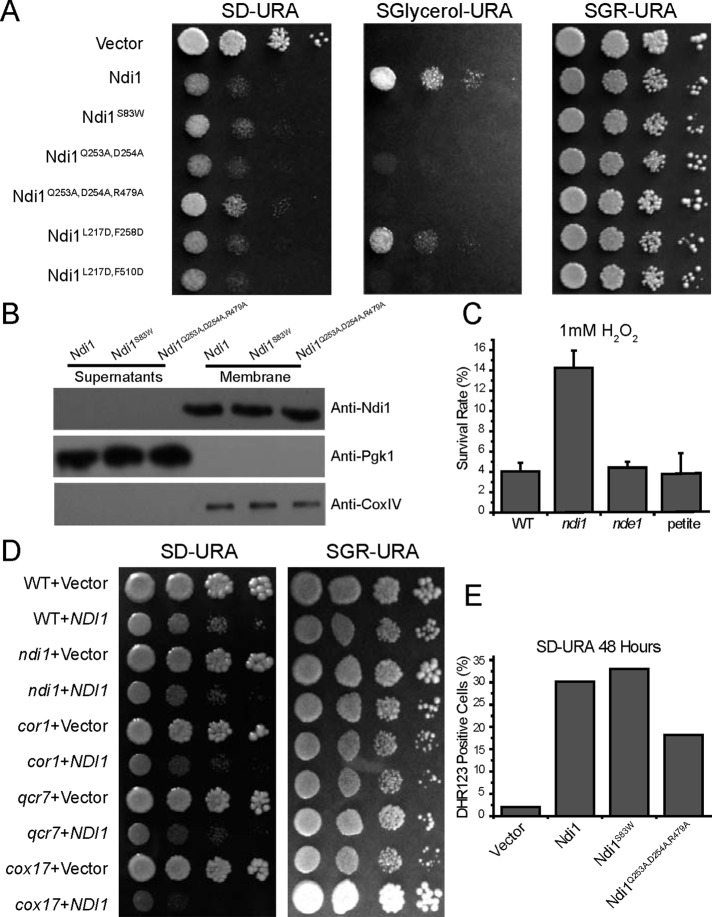
The mitochondrial electron transport chain (ETC) assimilates electrons from substrates such as NADH and polarizes the mitochondrial membrane potential for the ATP formation while serving as the major source of reactive oxygen species (ROS) production. In S. cerevisiae, Ndi1, together with Nde1—both mitochondrial ubiquinone oxidoreductases (internal or matrix side, and external or cytosolic side, respectively; Luttik et al., 1998; Small and McAlister-Henn, 1998)—acts as the major entry point of electrons into the respiratory chain. Previously we showed that when NDI1, but not NDE1, is overexpressed, yeast growth is strongly inhibited and apoptotic cell death is induced (Li et al., 2006). Conversely, ndi1-mutant cells have reduced ETC activity and are very resistant to oxidative stress. To determine whether the proapoptotic activity of Ndi1 is dependent on its ubiquinone oxidoreductase function, we made a series of point-mutated variants of Ndi1 (Figure 2A), which may have disrupted NADH dehydrogenase activity, based on structural and functional information (Feng et al., 2013), and tested the proapoptotic activities of these mutant forms. These Ndi1 variants were cloned into an ADH promoter-driven vector, transformed into an ndi1-deletion mutant, and analyzed for the loss of respiration activity. Some of these mutants indeed had lost the ubiquinone oxidoreductase function, as attested by their drastically reduced ability to grow on media with glycerol as the sole carbon source (Figure 2A). Spotting assays for several of these mutants, including Ndi1S83W, Ndi1Q253A,D254A, Ndi1Q253A,D254A,R479A, and Ndi1L217D,F510D, which had undetectable ubiquinone oxidoreductase function, and Ndi1L217D,F258D, which retained most ubiquinone oxidoreductase function, were further carried out to test their proapoptotic activity. All these mutants retained almost the same growth-inhibitory activity as the wild-type Ndi1 (Figure 2A). Several representative mutants were analyzed further for their protein expression and localization. All were shown to be expressed at roughly comparable levels and free in the cytosol, indicating that they are neither unstable nor mistargeted (Figure 2B).
FIGURE 2:
Proapoptotic activity of Ndi1 is independent of its ubiquinone oxidoreductase activity. (A) Loss of ubiquinone oxidoreductase activity is not associated with change of Ndi1 apoptotic activity. Shown here are spotting assays of ndi1-mutant strain transformed with the vector, Ndi1, Ndi1S83W, Ndi1Q253A,D254A, Ndi1Q253A,D254A,R479A, Ndi1L217D,F258D, and Ndi1L217D,F510D on SD-URA (respiration restricted), SGlycerol-URA (nonfermentable), and SGR-URA plates. Ndi1 overexpression inhibits cell growth in respiration-restricted media (glucose) but not glycerol or galactose/raffinose media. (B) Expression levels of several representative Ndi1 mutants are comparable to that of wild-type Ndi1, and these Ndi1 mutants are also associated with the mitochondrial membrane. (C, D) Ndi1 apoptotic activity is not dependent on ETC activity. (C) Viability testing of WT, ndi1, nde1, and petite yeast strains treated with 1 mM H2O2 indicates that loss of the ETC activity does not affect Ndi1-mediated apoptosis. (D) Various ETC mutants were tested for their effects on growth inhibition by Ndi1 overexpression. (E) Overexpression of Ndi1 mutants lacking oxidoreductase function also induced ROS when cultured in SD-URA, as shown by DHR123 staining with flow cytometry. Survival data are presented as the mean ±SEM of three independent experiments.
Having shown Ndi1-mediated apoptosis is independent of its NADH dehydrogenase function, we next explored whether it is independent of ETC activity in general. To determine whether a change of ETC would affect NDI1-mediated apoptosis, we tested the survival rates of wild-type, ndi1- and nde1-deletion mutant, and petite strains treated by H2O2. NDE1 is important for optimal cellular growth with a number of nonfermentable carbon sources, in particular ethanol, whereas petite yeast strain is defective in mitochondrial DNA and respiration. The nde1 and petite yeast strains almost had the same viability rates as the wild-type control. However, only ndi1-disrupted cells survived better, indicating that the Ndi1 proapoptotic property is not acting through ETC (Figure 2C). Furthermore, various ETC mutations in critical respiration genes such as cor1, qcr7, and cox17 did not abrogate the growth deficiency caused by NDI1 overexpression, again supporting the notion that Ndi1-mediated apoptosis is independent of ETC (Figure 2D). We previously reported that a majority but not a small minority of ETC mutants could affect NDI1-induced toxicity (Li et al., 2006). This discrepancy appears to arise from the instability of some of these mutant strains during strain storage and propagation. Aided by freshly generated mutants, we confirmed that the growth deficiency caused by NDI1 overexpression is independent of all ETC mutants tested. We therefore finally conclude that the proapoptotic activity of Ndi1 is distinguishable from its ubiquinone oxidoreductase function and is independent of mitochondrial ETC activity.
Ndi1-induced apoptosis is at least partially mediated by ROS production (Li et al., 2006). We next examined whether apoptotic cell death associated with Ndi1 mutants lacking oxidoreductase function is regulated by ROS. Using dihydrorhodamine 123 (DHR123) as an indicator, we showed that the overexpression of these Ndi1 mutants was still associated with ROS production (Figure 2E). To assess whether the ROS is functional to Ndi1-induced apoptosis, we transformed these mutant NDI1 into a sod2-mutant strain. Whereas normal-sized colonies were observed for the control vector, no colonies appeared on Ndi1 and Ndi1S83W plates 3 d after transformation, and only several small colonies appeared on plates transformed with Ndi1Q253A,D254A,R479A. This result indicates that ROS plays an important role both in the apoptotic cell death associated with Ndi1 and Ndi1 mutants lacking oxidoreductase function.
Mitochondrial Ndi1 translocates to the cytoplasm during apoptosis
In apoptosis, the mitochondria, cytoplasm, and nucleus closely interact with each other. The past 20 years of research has found that upon induction of apoptosis some mitochondrial factors escape. AIF, for example, translocates from the mitochondria into the nucleus. Conversely, some proteins translocate from the nucleus into the mitochondria or cytoplasm. Human Rad21 is cleaved and moves from the nucleus to the cytoplasm and acts as a nuclear signal for apoptosis (Chen et al., 2002; Pati et al., 2002), and Mcd1 is cleaved and translocated from the nucleus to the mitochondria to promote apoptosis (Yang et al., 2008).
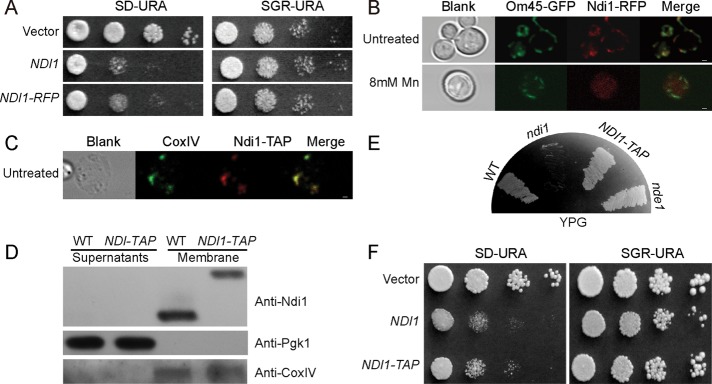
Considering the translocating behaviors of AIF during apoptosis, we examined Ndi1 localizations in the process of apoptosis. To determine whether the subcellular localization of Ndi1 may change when undergoing apoptosis, we used confocal microscopy to track the localization of Ndi1 in the cells. An NDI1-RFP fusion gene was constructed. By spotting assay, this red fluorescent protein (RFP)-tagged Ndi1 was shown to be properly functional; the ndi1-mutant strain overexpressing NDI1-RFP exhibited almost the same growth inhibition as that of the ndi1-mutant strain overexpressing wild-type NDI1, suggesting that NDI1-RFP retained its apoptotic activity (Figure 3A).
FIGURE 3:
Ndi1 translocates from mitochondria to cytoplasm during apoptosis. Ndi1-RFP is overexpressed, whereas Ndi1-TAP is under the endogenous regulation. (A) Ndi1-RFP maintains the proapoptotic function of Ndi1. (B) Ndi1-RFP is localized in the mitochondria under normal growth conditions but moves out during apoptosis. Om45-GFP is used to indicate mitochondria. Mn is used for apoptosis induction. (C, D) Ndi1-TAP colocalizes with mitochondrial inner membrane protein CoxIV, as detected by immunofluorescence and Western blot. (E) NDI1-TAP exhibited normal NADH dehydrogenase function like wild-type NDI1. It can use glycerol well. (F) Ndi1-TAP also shows similar apoptotic activity to the wild-type Ndi1. Bar, 1 μm.
The NDI1-RFP construct was then transformed into an Om45-GFP (BY4742) strain in which the mitochondrial Om45 (a major constituent of the mitochondrial outer membrane, located on the cytoplasmic face of the outer membrane) was fused with green fluorescent protein (GFP) to mark the mitochondria. The RFP and GFP signals can therefore be used to locate Ndi1 and mitochondria, respectively, when yeast is insulted by different apoptosis triggers. We used Mn as the apoptosis inducer. When treated with Mn, Ndi1-RFP relocalized from the mitochondria to the cytoplasm, whereas under no Mn stress Ndi1-RFP and Om45-GFP were perfectly colocalized (Figure 3B). This suggests that Ndi1 translocates from the mitochondria to the cytoplasm when stressed by apoptotic inducers.
A concern here is whether the big RFP tag or the Ndi1 overexpression might affect Ndi1’s behavior or subcellular localization—for example, whether these might alter the mitochondrial inner membrane residence of Ndi1. Because our homemade Ndi1 antibody does not react well with the N-truncated Ndi1, we tried a tandem affinity purification (TAP)–tagged, endogenously expressed NDI1-TAP strain in which Ndi1 is C-terminal fused to a TAP in its original chromosomal state (under endogenous promoter control). The fusion protein Ndi1-TAP is expressed in the mitochondria, as shown by both immunolocalization and Western blot (Figure 3, C and D). Although none of these experiments can pinpoint the inner membrane localization of Ndi1-TAP (like the normal Ndi1), its ability to grow on yeast extract/peptone/glycerol suggests that it is indeed correctly located (Figure 3E). This is because Ndi1 and Nde1 catalyze the internal and external NADH dehydrogenation, respectively. Loss of Ndi1 function results in poor growth in glycerol media, although growth is fine in ethanol media. The ability to use glycerol well indicates that Ndi1-TAP resides in the matrix side instead of the outer membrane. In addition, the ability to induce apoptosis is not affected by this TAP tagging (Figure 3F). All these demonstrate that Ndi1-TAP is functionally equivalent to native Ndi1. Using this NDI1-TAP strain, we also saw an obvious release of the protein to the cytoplasm (see Figure 4B)
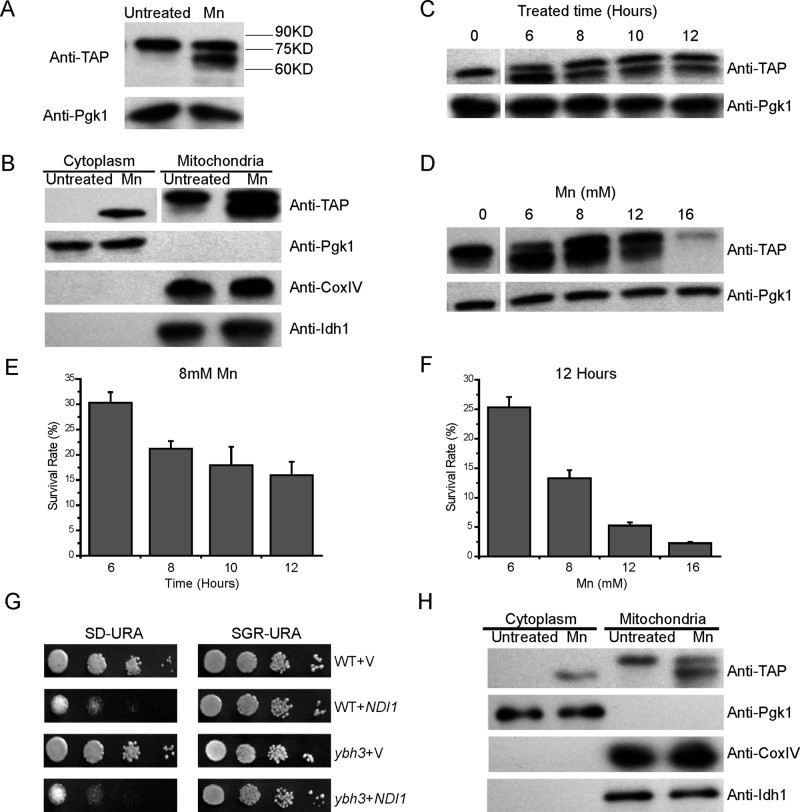
FIGURE 4:
Ndi1 is cleaved in mitochondria during apoptosis. (A) Ndi1 is cleaved when treated with Mn. NDI1-TAP yeast was treated or not treated with 8 mM Mn for 12 h, and the extracts were immunoblotted and probed with a TAP antibody. (B) Ndi1 cleavage occurs in the mitochondria. Both the full-length mature form of Ndi1 and the truncated Ndi1 were observed in the mitochondrial fraction, whereas only the truncated Ndi1 was found in the postmitochondrial fraction. Pgk1 served as the cytoplasm loading control, CoxIV as the mitochondrial loading control, and Idh1 as the matrix protein control. (C, D) The cleavage patterns of Ndi1 vs. treatment time and Mn concentration. (E, F) The survival rates of stressed NDI1-TAP yeast in C and D. (G, H) Ybh3 does not affect the apoptotic activity or release of Ndi1. (G) The proapoptotic activity of Ndi1 in the ybh3-deletion mutant tested by spotting assay. (H) The release of Ndi1 in NDI1-TAP,ybh3Δ background. Representative blots from at least three independent experiments are presented.
Ndi1 is N-terminal cleaved before its translocation out of the mitochondria
In the huge machinery network of apoptosis, many components are cleaved to be activated. A prominent case is the caspase family. Another crucial family is the B-cell lymphoma-2 family (Taylor et al., 2008). In both yeast and mammalian cells, some mitochondrial proteins involved in apoptosis were cleaved and then released from mitochondria. For example, during the process of apoptosis, AIF undergoes N-terminal proteolysis at the intermembrane space to form an apoptogenic fragment that can translocate into the nucleus (Otera et al., 2005). Considering that Ndi1 is a protein loosely bound to the mitochondrial inner membrane, we suspected that Ndi1 might also undergo some conformational or other kind of change to facilitate the translocation from mitochondria into cytoplasm.
To facilitate the detection of the potential degradation or cleavage of Ndi1, we used the NDI1-TAP strain. Indeed, besides the full-length Ndi1-TAP band, an additional, smaller band was also detected from the protein extract of yeast cells undergoing Mn-induced apoptosis, whereas under the normal growth condition this shorter band was not detectable (Figure 4A).
To check whether this cleavage occurs before or after the release from mitochondria, we used density gradient centrifugation to fractionate cell extracts into the mitochondrial and the cytoplasmic fractions, confirmed by respective antibodies against their endogenous marker proteins. It is obvious that during apoptosis, Ndi1 is cleaved before it moves out of the mitochondria, as evidenced by the observation that the postmitochondrial fraction only contained the shorter form of Ndi1 and the mitochondrial fraction contained both the full-length and shorter ones (Figure 4B). Another question is whether or not Ndi1 release from the matrix side is specific. It is possible that the entire inner membrane is permeabilized during Ndi1 release. As a control, we monitored the release of the tricarboxylic acid enzyme mitochondrial NAD(+)-dependent isocitrate dehydrogenase Idh1 during Ndi1 release (Cupp and McAlister-Henn, 1992). No Idh1 escaping to the cytoplasm was detected. Obvious truncation of Idh1 was not observed either (Figure 4B). Thus it appeared that the mitochondrial release of Ndi1 from the matrix side is at least relatively specific.
Further experiments showed that Ndi1 cleavage induced by Mn occurred in a time- and concentration-dependent manner (Figure 4, C and D). At very high levels of Mn (12 or 16 mM), an even greater percentage of yeast cells died (Figure 4, E and F), but less Ndi1 protein was converted into the shorter form, consistent with our previous finding that at high Mn levels yeast undergoes mostly necrosis rather than apoptosis. It appears that the cleavage of Ndi1 was specific in apoptosis.
Taken together, these data suggested that Ndi1 in its full length might be a target of some proteases that are activated during apoptosis induced by Mn, consequently generating a shorter fragment of Ndi1 to be released to the cytoplasm. Because the TAP signal is at the C-terminal of Ndi1, it is reasonable to consider that the smaller band observed in the Western analysis is Ndi1 with the very N-terminal removed.
The mitochondrial release of apoptotic proteins so far reported is normally from the intermembrane space. Considering the inner membrane location of Ndi1, it is not known how this translocation could occur. YBH3 was recently reported to be involved in mitochondrial outer membrane permeabilization (Büttner et al., 2011). We therefore tested whether mitochondrial release of Ndi1 and its apoptotic action would be affected in ybh3-deficient background. Neither Ndi1’s apoptotic function nor its release is obviously dependent on Ybh3 (Figure 4, G and H).
N-terminal cleavage is essential for Ndi1’s apoptotic action
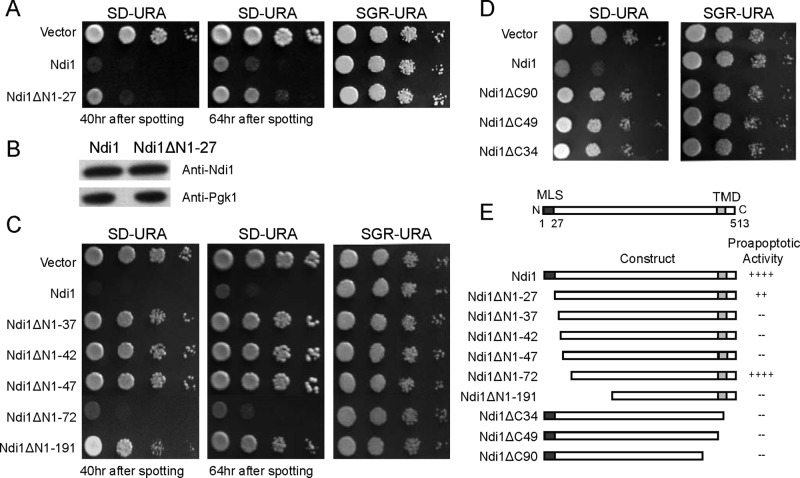
Is N-terminal removal functionally necessary for the proapoptotic activity of Ndi1? To address this question, we tested different forms of Ndi1 for their apoptotic activities. First, the mature full-length Ndi1 form without the mitochondrial signal peptide (cytosol-Ndi1 or Ndi1ΔN1-27) was expressed in the cytosol. Cytosol-Ndi1 showed much better growth than the yeast with the normal mitochondrial form of Ndi1. In other words, when the full-length mature form of Ndi1 was expressed in the cytosol, a much-reduced toxicity was observed than with the expression of its native mitochondrial form (Figure 5A). To exclude the possibility that this phenotype was caused by attenuated protein expression, we analyzed the protein level by Western blotting using a homemade murine Ndi1 antibody. No decrease of protein expression was observed (Figure 5B). It appears therefore that the cytoplasmic full-length Ndi1 would not be nearly as active in terms of executing Ndi1-mediated apoptosis.
FIGURE 5:
The N-terminal likely serves normally a protective role. (A) Cytosol-NDI1 (Ndi1ΔN1-27) displays much reduced toxicity as compared with normal NDI1. (B) A Western blotting control showing that cytosol-NDI1 was expressed similarly. (C) The N-terminal 40 or so amino acids of the mature Ndi1 are protective for Ndi1 toxicity. Without the first 45 (28–72) amino acids, Ndi1 (Ndi1ΔN1-72) becomes fully toxic when expressed in the cytoplasm. (D) The C-terminal of Ndi1 is required for Ndi1 toxicity. (E) A summary of the proapoptotic activities of various Ndi1 mutants. MLS, mitochondria localization signal; TMD, transmembrane domain; +, level of proapoptotic activity; –, no proapoptotic activity. Representative blot and spotting assay results from at least three independent experiments are presented.
Next we constructed a few N- and C-terminal–truncated Ndi1 variants (Figure 5E) and tested their apoptogenic activity. The truncated variants were designed according to potential protease cleavage sites, as well as functional and structural predictions. Several N-terminal–truncated Ndi1 variants were expressed in the cytoplasm to reproduce the growth-inhibitory activity of the native mitochondrial Ndi1. These variants include Ndi1ΔN1-37, Ndi1ΔN1-42, Ndi1ΔN1-47, Ndi1ΔN1-72, and Ndi1ΔN1-191. In particular, Ndi1ΔN1-72, which lacks the first 72 amino acids (or the potential mitochondrial localization sequence plus 45 amino acids in the N-terminal of the mature Ndi1), displayed almost the same inhibiting phenotype as the wild-type Ndi1 (Figure 5C). This form, which is predicted to be 4.6-kDa smaller than the wild-type mature Ndi1, closely matches the in vivo molecular weight of the shortened protein product detected by Western blotting during apoptosis. In contrast, removal of a few C-terminal residues reduced the cell growth–inhibition activity (Figure 5D). The proapoptotic activities of various Ndi1 forms are summarized in Figure 5E.
These data suggest that the N-terminal–cleaved Ndi1 is the activated form for apoptosis induction, and this cleavage involves about the first 40–50 amino acids of the N-terminal of the mature mitochondrial Ndi1.
DISCUSSION
To summarize briefly, besides displaying proapoptotic activity in respiration-repressed media and the aging process, yeast Ndi1 also participates in Mn- and H2O2-induced apoptosis. Ndi1-mediated apoptosis is not mediated by metacaspase, and the apoptogenic activity of Ndi1 is independent of its ubiquinone oxidoreductase function. When under stress, Ndi1 is cleaved at its N-terminal to be activated within mitochondria, and the truncated Ndi1 translocates to the cytoplasm to provoke apoptosis. To help visualization, Figure 6 is an illustration summarizing these processes: Ndi1 stops electron transferring when under stress, is cleaved, and moves out of the mitochondria to promote apoptosis.
FIGURE 6:
Model for the molecular mechanism of Ndi1’s dual actions. Ndi1 normally initiates electron transport from NADH to the electron transport chain. During stress, its normal function changes, and its N-terminal is cleaved. The cleaved Ndi1 then translocates from the mitochondria to the cytoplasm to provoke apoptosis.
A conserved and general yeast apoptotic factor
Ndi1 exhibits reasonable homology to mammalian AMID and AIF proteins, two players involved in apoptosis in mammalian cells. We found that yeast Ndi1 participates in a variety of stress-induced apoptosis, including that induced by aging, Mn, H2O2, and acetic acid. Therefore Ndi1 appears to be a proapoptotic factor broadly involved in yeast apoptosis. It has also been reported that AIF is involved in various cell death pathways, including lethal responses to N-methyl-d-aspartate and glutamate, DNA-damaging agents, oxidative stress, ceramide, hypoxia–ischemia, and growth factor deprivation (Joza et al., 2009), but the role of AMID has been less studied.
One protein with two separable functions
It had been reported that the oxidoreductase activity of AIF can be distinguished from its apoptotic function (Miramar et al., 2001). Here, we provided direct evidence that the proapoptotic activity of Ndi1 is independent of its original function in respiration. Therefore Ndi1 is a bifunctional molecule with redox and proapoptotic activities that are clearly distinct from each other. This observation is similar to those obtained from research on cytochrome c, which showed that its proapoptotic activity is independent of its redox status (Kluck et al., 1997, 2000). It is interesting to note that the Ndi1 homologue AMID in higher organisms appears to have lost the respiratory function; instead, the respiratory function is replaced by a sophisticated NADH dehydrogenase complex (complex I).
Because yeast Ndi1 shares the same oxidoreductase function as the higher eukaryotic multisubunit respiratory complex I (Marres et al., 1991; Velazquez and Pardo, 2001; Reinders et al., 2007), it is also called alternative complex I. More intriguingly, mammalian mitochondrial complex I protein was also found to play an important role in apoptosis. Complex I protein NDUFS3 is cleaved by granzyme A to initiate caspase-independent cell death (Martinvalet et al., 2008). It seems therefore that mammalian complex I and yeast Ndi1 share a similar function besides their roles in respiration. This makes sense from a biological point of view because when cells are under normal physiological conditions these proteins assimilate electrons to the ETC for ATP production. During apoptosis this respiration process is halted, and the proteins at the front of the respiration chains are converted to apoptotic proteins.
The protective role of the N-terminal
Ndi1 conversion from its role in respiration to its apoptotic role involves cleavage of its N-terminal. Therefore the N-terminal can be considered as an important switch and normally locks up the apoptotic activity and protects the cell from damage. This explains why the expression of Ndi1 with N-terminal in the cytosol has diminished cytotoxic activity, whereas the truncated form fully reproduces the native apoptotic effect.
N-terminal cleavage in mitochondria is triggered by stresses. The protease(s) involved in this process are not known. They might be of mitochondrial origin, as in the case of HtrA2. HtrA2 is synthesized as an inactive precursor and undergoes autoprocessing to become activated within the mitochondria when insulted by apoptotic stress (Martins et al., 2002). Chao et al. (2008) found that Hax1-mediated processing of HtrA2 by Parl (also a mitochondrial protease, presenilin associated and rhomboid-like) allows survival of lymphocytes and neurons. Alternatively, the responsible protease(s) can translocate into the mitochondria from other parts of the cell, as in the case of granzyme A, which translocates to the mitochondrial matrix to cleave the complex I protein NDUFS3 to initiate caspase-independent cell death (Martinvalet et al., 2008).
Translocation of Ndi1 during apoptosis
Protein relocalization during the execution of a specific assignment is not a novel phenomenon. Several kinds of molecules, such as cytochrome c (Liu et al., 1996; Eisenberg et al., 2007), EndoG (Li et al., 2001; Büttner et al., 2007), Smac/Diablo (Du et al., 2000), HtrA2/Omi (Suzuki et al., 2001; Fahrenkrog et al., 2004), and AIF, are released from the mitochondria at the beginning of apoptosis. As a protein located in the mitochondrial intermembrane space, cytochrome c escapes from the mitochondria and then facilitates the formation of the apoptosome, consisting of an adaptor (Apaf-1) and an initiator caspase (caspase-9), as well as ATP or 2′-deoxy ATP. EndoG is an apoptotic DNase-inducing nucleosomal fragmentation of DNA after release from the mitochondria. Smac is an inhibitor of X chromosome–linked inhibitor of apoptosis (XIAP), which releases from the mitochondria after the cells receive apoptotic stimuli. HtrA2 is a serine protease also released from the mitochondria and inhibits the function of XIAP by direct binding to it in a similar way to Smac. Finally, AIF is a flavoprotein that is normally confined to the mitochondria but translocates to the nucleus when apoptosis is induced. The translocation of Ndi1 from the mitochondria to the cytoplasm is confirmed by both immunoblotting and fluorescent confocal microscopy. However, the exact mechanistic role of Ndi1 after activation and translocation remains unclear, and further studies are needed to elucidate the cascade following the release of Ndi1 from the mitochondria.
The multiple and even redundant pathways to death
Our study also provides a glimpse into the connections among different components or pathways of yeast apoptosis. Ndi1 has an intimate connection with caspase-like activity induction, because in ndi1 mutants little caspase-like activity is observed even under various stresses. Two possibilities exist: either Ndi1 is required for the optimal induction of apoptosis-like activity directly, or caspase-like activity as observed is secondary to the initiation of apoptosis. In either case, although caspase activity helps the process of cell death, it is not absolutely required for it, indicating the existence of other, parallel participants downstream. Thus a likely cell death scenario is that when a normal yeast cell experiences a stress such as Mn, it turns on a program involving Yca1, Ndi1, caspase-like (Z-VAD binding) proteins, and some other players. All or at least some of them communicate with each other to work together to carry out cell death. Our current understanding indicates that both Ndi1 and Yca1 are connected with the so-far-unidentified caspase-like (Z-VAD binding) protein(s). Besides this caspase-like protein(s), other downstream players may work redundantly to ensure cell death. It therefore seems that when yeast, a relatively simple organism, decides to die, it activates multiple connected killers in a process that is not simple at all.
MATERIALS AND METHODS
Growth media and conditions, strains, and plasmids
Standard yeast media and growth conditions were used, except for those otherwise noted (Sherman, 1991). Experiments were usually performed in the background of BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), except that NDI1-TAP was in the background of S288C (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0; Open Biosystems, Huntsville, AL). The ndi1Δ (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ndi1::kanMX4) was obtained from Invitrogen (Carlsbad, CA). NDI1 variants were made by using NDI1 as the template and primers flanked by appropriate restriction sites. The PCR products were cloned into a yeast overexpression vector pADH-YES2, which is derived from pYES2 (Invitrogen) in which the GAL promoter was replaced by an ADH promoter. Yeast transformation was carried out by the standard lithium acetate method (Gietz and Schiestl, 1991). Empty vectors were also transformed in parallel to serve as negative controls. Single colonies of each transformed yeast strain were picked out and grown on uracil (URA)-dropout media (SD [2% glucose]-URA, SG [1% galactose]-R [1% raffinose]-URA, or SGlycerol [2%]-URA).
Spotting assays and viability tests
For spotting assays, yeast strains recovered from −80°C vials were shaken in SGR-URA liquid medium overnight for revitalization. Then they were adjusted to the identical OD600 of ∼0.5 and serially diluted to 10−1, 10−2, and 10−3. A 3-μl amount of each diluted yeast culture was spotted on SD-URA or other plates. SGR-URA plates served as the spotting control. Generally, pictures were taken after 2 or 3 d, depending on the growth.
For survival tests, yeast cells were shaken in test tubes and grown until they reached the exponential phase, and then different apoptotic inducers were added to induce apoptosis. After the cells were treated by Mn for 12 h or H2O2 for 100 min, the number of surviving colonies was determined by plating a small aliquot of the treated cultures on yeast extract/peptone/dextrose plates or selective plates (SD or SGR dropout plates).
In vivo staining of active metacaspase by flow cytometric analysis
A total of 5 × 106 cells were collected, washed with 1 ml of phosphate-buffered saline (PBS), resuspended in 200 μl PBS, and then incubated with FITC-VAD-fmk (Promega, Madison, WI) with a final concentration of 10 μM. For flow cytometric analysis, fluorescence-positive cells were counted using a flow cytometer (FACSVantageSE/DiVa; BD Biosciences, San Diego, CA) and EXP032 analysis software with excitation and emission wavelengths of 488 and 525 nm, respectively.
ROS detection by DHR123 staining
For the detection of accumulated ROS, DHR123 was incubated with yeast cells for 30 min and then analyzed by flow cytometry.
Membrane and postmembrane fractionation
We used a modified protocol as described by Yamashita et al. (2007). In brief, yeast cells were broken by three passages through a French pressure cell. Unbroken cells were removed by centrifugation at 4300 × g for 10 min. The supernatants were centrifuged at 250,000 × g for 60 min twice to spin down the membrane on one hand and to remove membrane contamination from the postmembrane fraction completely on the other. The membrane fraction was washed with PBS twice and dissolved in lysis buffer by sonication.
Immunoblotting
Immunoblotting experiments were performed mainly as described previously (Madeo et al., 2002). Samples were probed with the rabbit monoclonal antibody against TAP (Open Biosystems), mouse monoclonal antibody CoxIV (Invitrogen), goat polyclonal antibody Idh1 (Abnova, Taipei City, Taiwan), and a homemade murine polyclonal antibody against Ndi1 (recombinant yeast Ndi1 purified from Escherichia coli was used to immunize mice; 3 mo later the antiserum was collected, and the antibody was purified using protein G affinity chromatography [Invitrogen]).
Immunofluorescence
The standard protocol as described by Silver (2009) was applied. Specifically, the dilution ratio of CoxIV antibody was 1:200, and that of TAP antibody was 1:300. The secondary antibodies goat anti–rabbit-tetramethylrhodamine isothiocyanate and goat anti–mouse-FITC were diluted 200 times.
Confocal microscopy
For image acquisition, an Olympus FV1000 confocal microscope (Olympus, Tokyo, Japan) was used.
Fractionation of mitochondria and cytoplasm
Spheroblasts were prepared by digestion of yeast cells with lyticase as described previously (Jazwinski, 1990). Mitochondria preparation was modified according to Daum et al. (1982). After spinning down of the mitochondria by high-speed centrifugation, the supernatant was considered to be cytoplasm. The supernatant was centrifuged again to remove possible residual mitochondria. The mitochondrial part was washed by the isolation buffer to further reduce the cytoplasm components. Finally, the mitochondria were lysed by lysis buffer as used in immunoblotting.
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2011CB910900) and the National Natural Science Foundation of China (30871262, 30971568, and 31123004). We greatly appreciate the kind gift of purified recombinant Ndi1 protein from Yue Feng (Tsinghua University, Beijing, China). We thank Li Yu and Cong Yi (Tsinghua University, Beijing, China) for assistance with the confocal microscopy.
Abbreviations used:
- AIF
apoptosis-inducing factor
- AMID
AIF-homologous mitochondrion-associated inducer of death
- DHR123
dihydrorhodamine 123
- ETC
electron transport chain
- ROS
reactive oxygen species
- TAP
tandem affinity purification
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0281) on September 19, 2012.
The authors declare no conflict of interest.
REFERENCES
- Abdelwahid E, Rolland S, Teng X, Conradt B, Hardwick JM, White K. Mitochondrial involvement in cell death of non-mammalian eukaryotes. Biochim Biophys Acta. 2011;1813:597–607. doi: 10.1016/j.bbamcr.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner S, et al. Endonuclease G regulates budding yeast life and death. Mol Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Büttner S, et al. A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis. EMBO J. 2011;30:2779–2792. doi: 10.1038/emboj.2011.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Gutierrez D, Eisenberg T, Buttner S, Meisinger C, Kroemer G, Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- Chao JR, Parganas E, Boyd K, Hong CY, Opferman JT, Ihle JN. Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature. 2008;452:98–102. doi: 10.1038/nature06604. [DOI] [PubMed] [Google Scholar]
- Chen F, Kamradt M, Mulcahy M, ByuNY , Xu H, McKay MJ, Cryns VL. Caspase proteolysis of the cohesin component RAD21 promotes apoptosis. J Biol Chem. 2002;277:16775–16781. doi: 10.1074/jbc.M201322200. [DOI] [PubMed] [Google Scholar]
- Cupp JR, McAlister-Henn L. Cloning and characterization of the gene encoding the IDH1 subunit of NAD(+)-dependent isocitrate dehydrogenase from Saccharomyces cerevisiae. J Biol Chem. 1992;267:16417–16423. [PubMed] [Google Scholar]
- Daum G, Gasser SM, Schatz G. Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J Biol Chem. 1982;257:13075–13080. [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Eisenberg T, Buttner S, Kroemer G, Madeo F. The mitochondrial pathway in yeast apoptosis. Apoptosis. 2007;12:1011–1023. doi: 10.1007/s10495-007-0758-0. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Sauder U, Aebi U. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J Cell Sci. 2004;117:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- Feng Y, et al. Structural insight into the type-II mitochondrial NADH dehydrogenases. Nature. 2013 doi: 10.1038/nature11541. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Greenwood MT, Ludovico P. Expressing and functional analysis of mammalian apoptotic regulators in yeast. Cell Death Differ. 2010;17:737–745. doi: 10.1038/cdd.2009.177. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. Preparation of extracts from yeast. Methods Enzymol. 1990;182:154–174. doi: 10.1016/0076-6879(90)82015-t. [DOI] [PubMed] [Google Scholar]
- Joza N, Pospisilik JA, Hangen E, Hanada T, Modjtahedi N, Penninger JM, Kroemer G. AIF: not just an apoptosis-inducing factor. Ann NY Acad Sci. 2009;1171:2–11. doi: 10.1111/j.1749-6632.2009.04681.x. [DOI] [PubMed] [Google Scholar]
- Kluck RM, Ellerby LM, Ellerby HM, Naiem S, Yaffe MP, Margoliash E, Bredesen D, Mauk AG, Sherman F, Newmeyer DD. Determinants of cytochrome c pro-apoptotic activity. The role of lysine 72 trimethylation. J Biol Chem. 2000;275:16127–16133. doi: 10.1074/jbc.275.21.16127. [DOI] [PubMed] [Google Scholar]
- Kluck RM, Martin SJ, Hoffman BM, Zhou JS, Green DR, Newmeyer DD. Cytochrome c activation of CPP32-like proteolysis plays a critical role in a Xenopus cell-free apoptosis system. EMBO J. 1997;16:4639–4649. doi: 10.1093/emboj/16.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Li W, Sun L, Liang Q, Wang J, Mo W, Zhou B. Yeast AMID homologue Ndi1p displays respiration-restricted apoptotic activity and is involved in chronological aging. Mol Biol Cell. 2006;17:1802–1811. doi: 10.1091/mbc.E05-04-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Zhou B. Copper and manganese induce yeast apoptosis via different pathways. Mol Biol Cell. 2007;18:4741–4749. doi: 10.1091/mbc.E07-05-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Ludovico P, Sousa MJ, Silva MT, Leao C, Corte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology. 2001;147:2409–2415. doi: 10.1099/00221287-147-9-2409. [DOI] [PubMed] [Google Scholar]
- Luttik MA, Overkamp KM, Kotter P, de Vries S, van Dijken JP, Pronk JT. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem. 1998;273:24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Frohlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Frohlich E, Ligr M, Grey M, Sigrist SJ, Wolf DH, Frohlich KU. Oxygen stress: a regulator of apoptosis in yeast. J Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, et al. A caspase-related protease regulates apoptosis in yeast. Mol Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- Marres CA, de Vries S, Grivell LA. Isolation and inactivation of the nuclear gene encoding the rotenone-insensitive internal NADH: ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur J Biochem. 1991;195:857–862. doi: 10.1111/j.1432-1033.1991.tb15775.x. [DOI] [PubMed] [Google Scholar]
- Martins LM, et al. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J Biol Chem. 2002;277:439–444. doi: 10.1074/jbc.M109784200. [DOI] [PubMed] [Google Scholar]
- Martinvalet D, Dykxhoorn DM, Ferrini R, Lieberman J. Granzyme A cleaves a mitochondrial complex I protein to initiate caspase-independent cell death. Cell. 2008;133:681–692. doi: 10.1016/j.cell.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramar MD, Costantini P, Ravagnan L, Saraiva LM, Haouzi D, Brothers G, Penninger JM, Peleato ML, Kroemer G, Susin SA. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J Biol Chem. 2001;276:16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- Otera H, Ohsakaya S, Nagaura Z, Ishihara N, Mihara K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 2005;24:1375–1386. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati D, Zhang N, Plon SE. Linking sister chromatid cohesion and apoptosis: role of Rad21. Mol Cell Biol. 2002;22:8267–8277. doi: 10.1128/MCB.22.23.8267-8277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wagner K, Zahedi RP, Stojanovski D, Eyrich B, van der Laan M, Rehling P, Sickmann A, Pfanner N, Meisinger C. Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol Cell Proteomics. 2007;6:1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Silver P. Indirect immunofluorescence labeling in the yeast Saccharomyces cerevisiae. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5317. pdb.prot5317. [DOI] [PubMed] [Google Scholar]
- Small WC, McAlister-Henn L. Identification of a cytosolically directed NADH dehydrogenase in mitochondria of Saccharomyces cerevisiae. J Bacteriol. 1998;180:4051–4055. doi: 10.1128/jb.180.16.4051-4055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- Velazquez I, Pardo JP. Kinetic characterization of the rotenone-insensitive internal NADH: ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Arch Biochem Biophys. 2001;389:7–14. doi: 10.1006/abbi.2001.2293. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Lam E. Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J Biol Chem. 2005;280:14691–14699. doi: 10.1074/jbc.M413527200. [DOI] [PubMed] [Google Scholar]
- Wissing S, et al. An AIF orthologue regulates apoptosis in yeast. J Cell Biol. 2004;166:969–974. doi: 10.1083/jcb.200404138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Xu LG, Li X, Zhai Z, Shu HB. AMID, an apoptosis-inducing factor-homologous mitochondrion-associated protein, induces caspase-independent apoptosis. J Biol Chem. 2002;277:25617–25623. doi: 10.1074/jbc.M202285200. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Nakamaru-Ogiso E, Miyoshi H, Matsuno-Yagi A, Yagi T. Roles of bound quinone in the single subunit NADH-quinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae. J Biol Chem. 2007;282:6012–6020. doi: 10.1074/jbc.M610646200. [DOI] [PubMed] [Google Scholar]
- Yang H, Ren Q, Zhang Z. Cleavage of Mcd1 by caspase-like protease Esp1 promotes apoptosis in budding yeast. Mol Biol Cell. 2008;19:2127–2134. doi: 10.1091/mbc.E07-11-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]