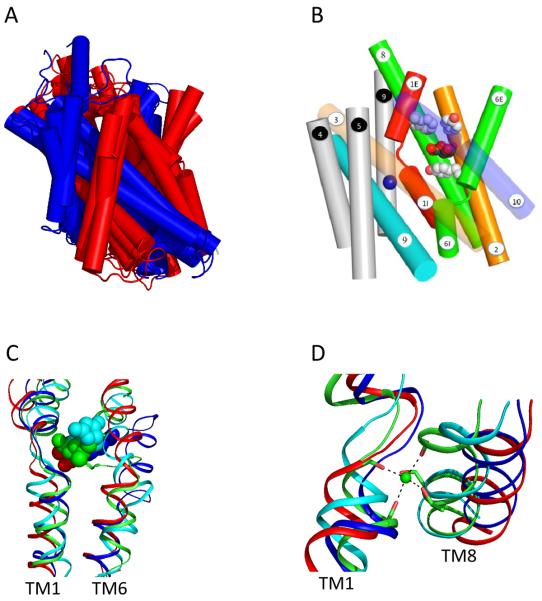
Figure 3. The location of substrate and the common sodium binding site.
A) Structural alignment of the core domains from vSGLT, BetP, LeuT and Mhp1 yield an RMSD between 3.8 Å and 4.5 Å. TM1-TM5 and TM6-TM10 are colored red and blue respectively. B) Side view of vSGLT showing the location of the sugar binding site, the extracellular (M73, Y87 and F424) and intracellular (Y263) gates. The numbering of the TMs is according to the convention shown in Figure 1. C) A structural alignment of TM1, TM6 and the substrate-binding site for vSGLT (red), BetP (blue), LeuT (green) and Mhp1 (cyan). D) A structural alignment of TM1, TM8 and the conserved sodium-binding site for vSGLT (red), BetP (blue), LeuT (green) and Mhp1 (cyan).