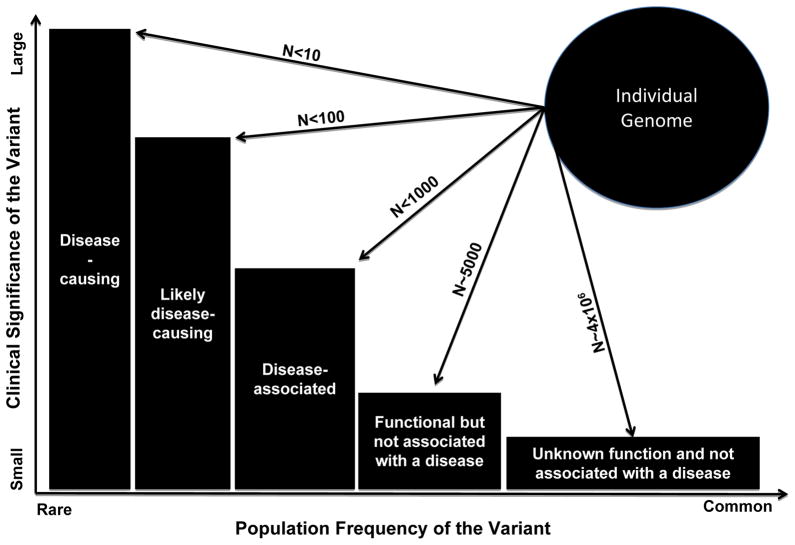
Figure 1. Plot showing population frequency of the DNA sequence variants (DSVs), abundance in an individual genome and the clinical significance (effect size) of the variants.
Rare alleles impart larger effect sizes and in general, have a greater clinical significance than common alleles, which have modest effect sizes and typically not clinically significant. Each genome contains a few disease-causing variants, a category for which there is strong genetic evidence of causality, typically through studies in large pedigrees with single gene-disorders. Each genome also contains a number rare variants with large effect sizes that are considered “Likely disease-causing variants” as the genetic evidence for their causal role is not sufficiently conclusive. This group typically includes rare variants that are identified in the affected individuals, wherein co-segregation with the phenotype cannot be robustly established because or small size of the families or incomplete penetrance. Disease-associated variants are relatively more common and are those that in large –scale studies, such as GWAS have been associated with the phenotype. However, they are neither sufficient to cause the disease nor necessary but rather increase the risk of the disease. Each genome/exome has about 7,000 non-synonymous putatively functional variants. The most abundant variants encompasses about 4 million DSV that neither have a known biological function or are associated with a phenotype.