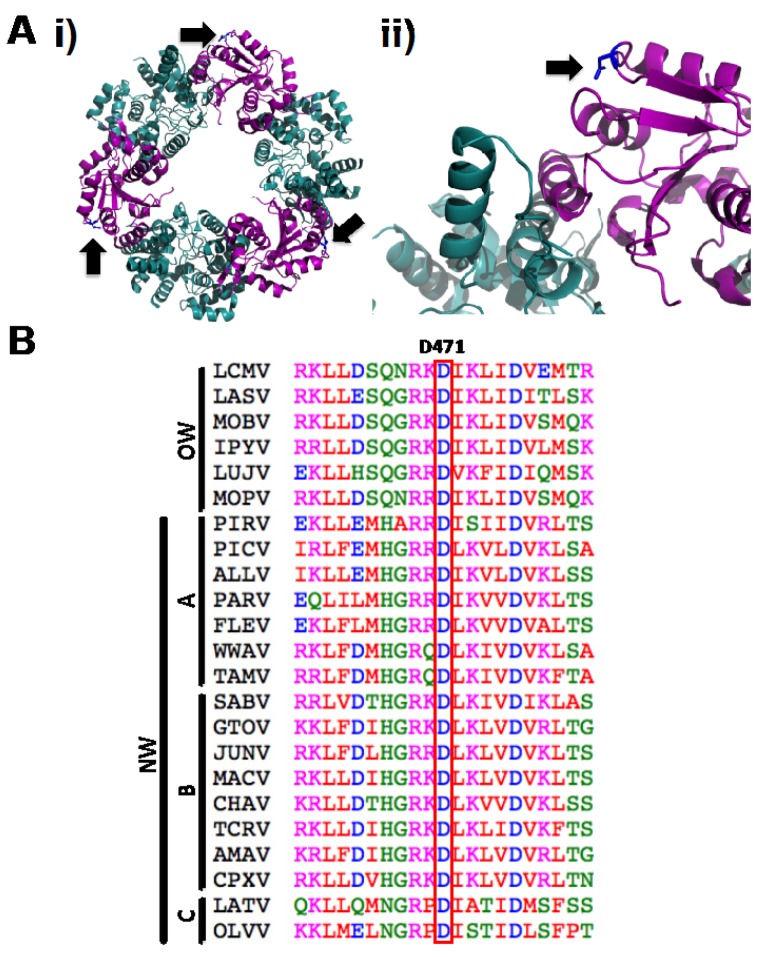
Figure 9.
Structural extrapolation and sequence alignment analysis of LCMV-NP D471 residue. (A) Cartoon diagrams of LASV-NP crystal structure [29] indicating localization of amino acid residue D471. (i) Residue D471 in the trimeric structure, indicated by black arrows. (ii) N-C interface of the LASV-NP structure, where the side chain of residue D471 is indicated in blue and pointed by a black arrow. N-terminal and C-terminal regions of LASV-NP are indicated in cyan and magenta, respectively. (B) D471 amino acid residue is highly conserved between arenavirus NPs. Amino acid sequence alignment of arenavirus NPs of the region spanning D471 residue. Representative members of Old World (OW) and the New World (NW) clades A, B and C arenaviruses were included. LCMV, Lymphocytic choriomeningitis virus (AY847350.1); LASV, Lassa virus (HQ688673.1); MOBV, Mobala virus (AY342390.1); IPYV, Ippy virus (NC_007905.1); LUJV, Lujo virus (JX017360.1); MOPV, Mopeia virus (NC_006574.1); PIRV, Pirital virus (NC_005894); PICV, Pichinde virus (AF081553.1); ALLV, Allpahuayo virus (NC_010253.1); PARV, Parana virus (AF512829.1); FLEV, Flexal virus (NC_010757.1); WWAV, White water arroyo virus (EU486820.1); TAMV, Tamiami virus (EU486821.1); SABV, Sabia virus (NC_006317.1); GTOV, Guanarito virus (NC_005077.1); JUNV, Junin virus (JN801476.1); MACV, Machupo virus (NC_005078.1); CHAV, Chapare virus (NC_010562.1); TCRV, Tacaribe virus (NC_004293.1); AMAV, Amapari virus (NC_010247.1); CPXV, Cupixi virus (NC_010254.1), LATV, Latino virus (AF512830.1); OLVV, Oliveros virus (NC_010248.1). ClustalW2 program was used for alignment [57]. Amino acid are colored according to chemical properties: red (hydrophobic and aromatic amino acids), blue (acidic), magenta (basic), green (hydroxyl and amine containing), as specified by the EMBL-EBI CLUSTALW 2.0.8 multiple sequence alignment program.