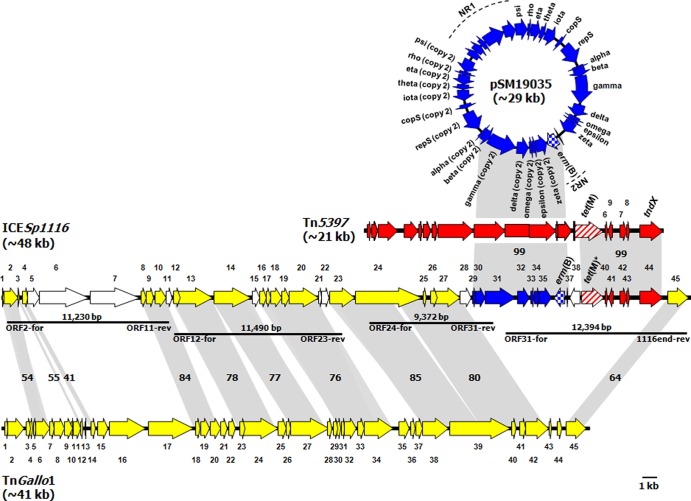
Fig 1.
ORF map and genetic organization of ICESp1116 from S. pyogenes strain A-3 (accession no. HE802677) and its alignment with the ORF maps of S. gallolyticus transposon TnGallo1 (accession no. FN597254), S. pyogenes plasmid pSM19035 (accession no. AY357120), and C. difficile transposon Tn5397 (accession no. AF333235). The ORFs, indicated as arrows pointing in the direction of transcription, are numbered consecutively in ICESp1116 (orf1 to orf45, with predicted functions reported in Table S1 in the supplemental material) and in TnGallo1 (orf1 to orf45). In pSM19035, where the two nonrepeated sequences NR1 and NR2 are marked by broken lines, the ORFs are numbered according to the specific designations reported in GenBank (the designations of the NR1 ORFs are not indicated). In Tn5397, the ORFs are also numbered according to the specific designations reported in GenBank [those upstream of tet(M) are not indicated]. TnGallo1 ORFs and related ORFs of ICESp1116 are depicted as yellow arrows. pSM19035 ORFs and ICESp1116 ORFs of the pSM19035 fragment are depicted as blue arrows [with erm(B) checkered]. Tn5397 ORFs and ICESp1116 ORFs of the Tn5397 fragment are depicted as red arrows [with tet(M) striped]. The other ICESp1116 ORFs are depicted as white arrows. Gray areas between ORF maps denote amino acid identities as reported (%). Horizontal bars indicate the amplicons (each with the relevant primer pair and size) allowing PCR mapping.