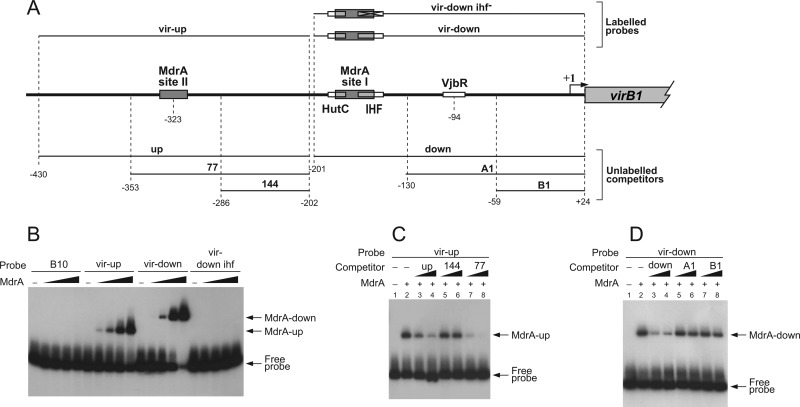
Fig 3.
Analysis of the interaction between MdrA and the virB promoter. (A) Schematic representation of genomic sequences corresponding to the regulatory region of the virB operon, radiolabeled probes used for EMSA, or unlabeled competitors. Positions relative to the transcription start site are indicated. MdrA-, HutC-, IHF-, and VjbR-binding sites are indicated as boxes. The diagonally crossed box indicates the nonrelated sequences that replace the IHF-binding site in probe vir-down-ihf−. (B) EMSA performed with the indicated probes and 0, 10, 20, 40, or 60 nM MdrA. (C) EMSA performed with probe vir-up, MdrA, and the indicated unlabeled DNA fragments as competitors. Protein concentrations were as follows: line 1, no protein; lines 2 to 8, 30 nM. Concentrations of unlabeled DNA competitors were as follows: lanes 1 and 2, no competitor DNA; lanes 3, 5, and 7, 150 ng; lanes 4, 6, and 8, 360 ng. (D) EMSA performed with probe vir-down, MdrA, and the indicated unlabeled DNA fragments as competitors. The concentrations of protein and unlabeled DNA competitors were the same as for panel C.