Abstract
The ferric uptake regulator (Fur) protein has been shown to function as a repressor of transcription in a number of diverse microorganisms. However, recent studies have established that Fur can function at a global level as both an activator and a repressor of transcription through both direct and indirect mechanisms. Fur-mediated indirect activation occurs via the repression of additional repressor proteins, or small regulatory RNAs, thereby activating transcription of a previously silent gene. Fur mediates direct activation through binding of Fur to the promoter regions of genes. Whereas the repressive mechanism of Fur has been thoroughly investigated, emerging studies on direct and indirect Fur-mediated activation mechanisms have revealed novel global regulatory circuits.
INTRODUCTION
Iron homeostasis is a highly regulated process in bacteria, as iron both is an essential nutrient and when in excess, can lead to toxicity via the production of hydroxyl or peroxide radicals. To maintain iron homeostasis, many bacteria utilize iron-binding transcriptional regulators, which, upon binding to free iron, are triggered to regulate transcription of genes involved in maintaining intracellular iron levels. Iron-binding proteins are members of a large class of metal ion-binding transcriptional regulators, includin MntR, DtxR, and Zur (58). These regulators act as environmental sensors of essential metals, including iron, and modulate gene expression accordingly. The master regulator of iron homeostasis is an iron-binding transcription factor termed the Ferric Uptake Regulator (Fur).
The first indication that bacterial iron homeostasis relied on a single central regulator was published by Ernst et al. in 1978 (31). Those studies showed that a Salmonella enterica serovar Typhimurium mutant lacked iron-responsive regulation of many genes, including those involved in iron-enterochelin and ferrochrome uptake. That mutant was termed an iron (Fe) uptake regulation (fur) mutant (31). A similar mutant was soon isolated in Escherichia coli, and the E. coli fur gene was subsequently cloned and sequenced (45, 84). Orthologues of fur have since been identified in numerous Gram-negative and Gram-positive species, and it has been shown that Fur proteins share a high degree of sequence homology between species (Fig. 1). In most organisms, Fur is present as a 15-to-17-kDa protein that forms dimers in the presence of iron (II) or other divalent cations (6, 24, 74, 91). As revealed by analysis of several crystal structures of the Fur protein from various pathogens, the amino terminus of Fur has been shown to bind to DNA whereas the carboxyl terminus is involved in dimer formation (26, 81, 91). Analysis of the crystal structure of Fur has also identified multiple metal-binding sites. These metal-binding sites contain a conserved histidine-histidine-aspartic acid-histidine (HHDH) motif (a specific region shown to be involved in cofactor binding), and mutagenesis studies have shown these four amino acids are crucial for Fur function (59, 83). As a repressor, the iron-bound Fur dimer binds to the −10 and −35 promoter regions to exclude binding of RNA polymerase, which results in the inhibition of transcriptional initiation (32) (Fig. 2A). The DNA sequence recognized by repressive Fur (designated a Fur box) was initially defined as a conserved 19-bp sequence, GATAATGATAATCATTATC, in E. coli (24). Fur boxes determined in other Gram-positive or Gram-negative bacteria are similar to this consensus sequence (7, 24, 27, 37, 44, 69, 78, 80, 96, 98, 104, 105) (Table 1). Subsequently, the Fur box was interpreted as 9-1-9 inverted repeats (GATAATGAT-A-ATCATTATC), or a hexameric repetition of nATwAT (24, 33). The Fur boxes in Bacillus subtilis and Helicobacter pylori are represented by shorter inverted repeats of 7-1-7 (TGATAATnATTATCA and TAATAATnATTATTA, respectively) (37, 80). Based on these studies and the crystal structure of Fur, it is predicted that two Fur dimers simultaneously bind one Fur box and the two Fur dimers may occupy three to four hexameric repeats (81). This consensus sequence has been used to successfully predict novel Fur-regulated genes in silico (27, 44, 53).
Fig 1.
Alignment of Fur orthologues from both Gram-negative and Gram-positive bacteria.
Fig 2.
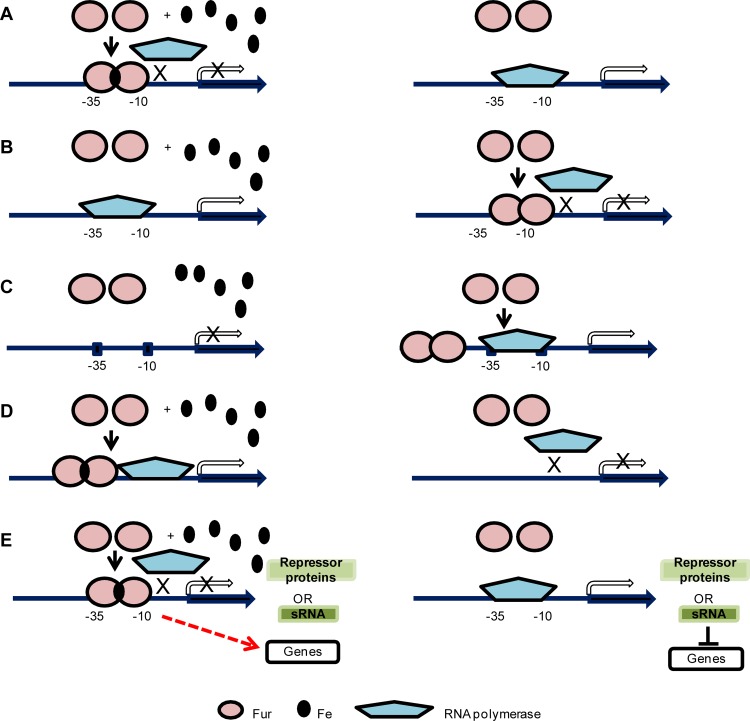
Mechanisms of Fur-mediated regulation. (A) Iron-bound Fur dimer binds to the −10 and −35 motifs in the promoter region, blocking binding of RNA polymerase and thus reducing transcription (left panel). Without iron, Fur does not bind to the promoter region, thereby allowing transcription via RNA polymerase (right panel). This mechanism has been well established by experimental evidence (32). (B) In the apo-Fur repression mechanism, when iron is present, Fur does not bind to the promoter region, resulting in transcription of the gene by RNA polymerase (left panel). Without iron, the apo-Fur dimer binds to the −10 and −35 motifs in the promoter region to inhibit binding of RNA polymerase and repress transcription (right panel). Apo-Fur repression was experimentally demonstrated in H. pylori (13, 72). (C) In the apo-Fur activation mechanism, Fur does not bind to the promoter region; thus, gene transcription is not active when iron is present (left panel). Without iron, the apo-Fur dimer binds to a site further upstream of the −10 and −35 motifs in the promoter region and upregulates transcription (right panel). This mechanism was first described in V. vulnificus (57). (D) Iron-bound Fur dimer binds to the promoter region and upregulates transcription (left panel). Without iron, Fur does not bind to the promoter region to initiate transcription (right panel). A few cases have been reported that indicate Fur-mediated direct activation through binding to defined promoter regions (50, 77, 105). (E) Iron-bound Fur dimer binds to −10 and −35 motifs in the promoter region and represses a negative regulator such as a protein repressor or a small RNA. Subsequently, genes that are repressed by the negative regulators are then transcribed and these genes show indirect Fur activation (left panel). Without iron and Fur repression, the negative regulators are transcribed and repress their target genes (right panel). Fur-repressed small RNAs have been reported in E. coli (61–64), V. cholerae (21), P. aeruginosa (103), and B. subtilis (38) as well as in the two pathogenic Neisseria species (28, 65, 66). Known Fur-repressed protein regulators include H-NS in S. enterica serovar Typhimurium (95) and MpeR in N. gonorrhoeae (53).
Table 1.
Fur box consensus sequences of various Gram-positive or Gram-negative bacteria
| Species | Consensus sequence of Fur boxa | Reference(s) |
|---|---|---|
| Bacillus subtilis | TGATAATnATTATCA | 7, 37 |
| Staphylococcus aureus | GTTCATGATAATCATTATC | 104 |
| Escherichia coli | GATAATGATAATCATTATC | 23 |
| Pseudomonas aeruginosa | GATAATGATAATCATTATC | 78 |
| Salmonella enterica serovar Typhimurium | GATAATGATAATCATTATC | 96 |
| Vibrio cholerae | GATAATGATAATCATTATC | 69 |
| Helicobacter pylori | TAATAATnATTATTA | 80, 98 |
| Neisseria meningitidis | nATwATnATwATnATwATn | 44 |
| Neisseria gonorrhoeae | T-ATAAT-ATTATCA | 27, 105 |
See text for explanation of the significance of the hyphens and the uppercase and lowercase characters in the sequences.
Fur FUNCTIONS AS A GLOBAL REGULATORY PROTEIN
Recently, global analyses of iron- and/or Fur-responsive transcriptomes of diverse bacterial pathogens, such as H. pylori, Pseudomonas syringae, Vibrio cholerae, Yersinia pestis, Haemophilus influenzae, S. enterica serovar Typhimurium, and Listeria monocytogenes, have revealed a number of novel regulatory roles for Fur (12, 20, 40, 56, 68, 94, 101, 102, 107). First, Fur has been demonstrated to repress transcription even in the absence of iron, a process termed apo-Fur regulation (13) (Fig. 2B). apo-Fur repression has been primarily characterized in H. pylori and has not yet been well described in other bacteria (13, 72). Although iron may not be important for apo-Fur function, it is possible that other metal ions play a role in apo-Fur-mediated transcriptional control (24, 74, 91). Second, in addition to its role as a repressor, Fur can also function as an activator in both the iron-bound form and apo form. In V. vulnificus, apo-Fur-mediated activation was shown to positively regulate the fur gene itself (57) (Fig. 2C). Transcriptome analysis has also identified additional genes that are activated by iron-bound Fur (12, 56, 68, 101, 102, 107).
Theoretically, transcriptional activation through Fur can be fulfilled by several pathways, including both direct (Fig. 2D) and indirect (Fig. 2E) mechanisms. A few examples of Fur-mediated indirect activation have been reported. Fur may activate transcription indirectly via repression of a small RNA (sRNA), such as RyhB of E. coli (5, 62, 99). As a result, the repressed targets of RyhB, consisting of sdhCDAB, acnA, fumA, and sodB, are activated when Fur is present (61–64). Fur-repressed sRNAs and their target genes have been identified in the Fur regulons of other bacteria, such as RyhB of V. cholerae (21), PrrF1 and PrrF2 of P. aeruginosa (103), and FsrA of B. subtilis (38), as well as NrrF in Neisseria meningitidis (65, 66). In addition to sRNAs, Fur may repress a proteinaceous repressor to activate downstream genes. For example, transcriptional activation of hilA in S. enterica serovar Typhimurium was demonstrated to result from the direct repression of a negative regulator of hilA, H-NS (histone-like nucleotide binding protein), by Fur (95). Fur has also been shown to directly bind to the promoter regions of Fur-activated genes. In E. coli, Fur and H-NS compete for overlapping binding sites within the promoter regions of ftnA, resulting in derepression of ftnA transcription (77). The transcription of the V. cholerae porin ompT gene was positively regulated by iron-bound Fur through the direct binding of Fur to the promoter region (17). However, in contrast to the wealth of studies describing Fur-mediated repression and despite the few previous examples, Fur-mediated direct activation via binding to defined promoter regions has been less studied.
In the remainder of this review, we discuss recent studies of Fur-mediated global regulatory circuits in the pathogenic Neisseria (N. meningitidis and N. gonorrhoeae). Since genomic analysis has revealed that there are fewer than 60 predicted regulatory proteins in the Neisseria genomes compared to ∼200 in the E. coli genome (85), we propose that Fur-mediated regulation in these organisms may have more global and versatile consequences for gene expression and associated pathogenic mechanisms.
Fur REGULON OF N. MENINGITIDIS
An N. meningitidis global microarray analysis has identified 233 genes whose transcription levels were affected by growth under iron-replete versus -depleted conditions (44). Of these 233 genes, ∼ 50% were predicted by in silico analysis to contain Fur boxes in their promoter regions (44). In addition, the majority of the predicted Fur binding promoter regions were experimentally demonstrated to bind Fur in vitro (44). A subsequent study examining iron regulation in a meningococcal fur mutant strain identified 83 genes whose iron-responsive regulation required Fur (22). Interestingly, 44 of those genes were repressed and 38 were activated, defining a new role for Neisseria Fur in activation of gene expression (22).
As demonstrated by electrophoretic mobility shift assay (EMSA) and/or DNase I footprinting results, genes and operons directly repressed by Fur in N. meningitidis encode proteins which can be classified into four major groups based on their functions: iron uptake and transport, energy metabolism and biosynthesis, toxin and stress responses, and regulation (Table 2) (22, 24, 44, 65, 66, 90). As expected, genes encoding iron uptake and transport proteins such as tbpA, tbpB, lbpA, and lbpB are repressed by Fur under iron-replete conditions, in agreement with the primary role of Fur as a maintainer of iron homeostasis (Table 2). The second group of Fur-repressed genes includes a large number of genes involved in energy metabolism and biosynthesis (Table 2). The protein products of these genes appear to enable bacterial growth, whereas their roles in pathogenesis have not yet been investigated. Interestingly, these genes have few homologs in N. gonorrhoeae (Table 2). The third group includes genes involved in virulence and bacterial adaption. It has been shown that FrpC-like proteins of N. meningitidis may play a role in pathogenesis (35, 79), and several frpA- and frpC-related gene loci, including NMB0364, NMB0584, NMB1405, and NMB1412 to -1414, were repressed by Fur directly (Table 2) (44). Several chaperone proteins and putative transposases, in addition to RecN, which are involved in DNA recombination and repair processes are also proposed to support bacterial adaption to the host environment and are directly repressed by Fur (Table 2) (44). The last group of Fur-repressed genes can be classified as regulators. A large percentage of Fur-dependent genes did not appear to be directly regulated by Fur, as demonstrated by the inability of Fur to bind to the promoter regions, which suggests the involvement of secondary regulators (22, 44). So far, only the small RNA NrrF has been identified as a transcriptional regulator directly controlled by Fur in N. meningitidis (Table 2) (65, 66). It is therefore logical to predict that additional, as-yet-uncharacterized Fur-controlled regulators could exist. Conversely, nine genes or operons directly activated by Fur fall into two major groups: iron storage and oxidative stress resistance genes and gene loci such as sodB, kat, norB, aniA, and NMB1438 to -1436 and energy metabolism loci such as the nuo complex (NMB0242 to -0244) (Table 2) (22, 44). In addition, a large number of hypothetical proteins under Fur regulation await further investigation (22, 44).
Table 2.
Genes directly regulated by Fur in N. meningitidis and N. gonorrhoeaea
| Category |
N. meningitidis MC58 |
N. gonorrhoeae FA1090 |
||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Function | Expt(s) | Reference(s) | Gene | Function | Expt(s) | Reference(s) | |
| Direct repression | ||||||||
| Iron acquisition | NMB0205 | fur, ferric uptake regulator protein | EM, FP | 44 | NGO1779 | fur | EM, FT | 53, 86 |
| NMB0634 | fbpA, iron binding protein | EM | 44 | NGO0215 to -0217 | fbpABC | EM, FP, FT | 25, 36, 53 | |
| NMB1668 | hmbR, hemoglobin receptor | FP | 22 | NGO1318 | hemO-hemR, heme utilization protein | EM, FT | 53, 86 | |
| NMB0460 | tbp2, transferrin binding protein B | EM | 44 | NGO1496 | tbpB | FT | 53 | |
| NMB0461 | tbp1, transferrin binding protein A | EM | 44 | NGO1495 | tbpA | FT | 53 | |
| NMB1540 | lbpA, lactoferrin binding protein A | EM | 44 | lbpAb | Pc | 9, 41 | ||
| NMB1541 | lbpB, lactoferrin binding protein B | EM, FP | 22, 44 | lbpBb | P | 8 | ||
| NMB1730 | tonB, energy transducer | FP | 22 | NGO2176 | tonB | EM | 86 | |
| NMB1728 | exbD, biopolymer transport protein | EM | 44 | |||||
| NMB0175 | zupT, zinc transporter | FP | 22 | |||||
| NMB1988 | frpB (fetA), ferric enterobactin receptor | EM, FP | 22, 44 | NGO2093 | fetA | FT | 53 | |
| NGO2092 | fetB, ferric enterobactin periplasmic binding protein | EM, FT | 53 | |||||
| NGO0024 | putative FetB2 protein | FT | 53 | |||||
| NGO0553 | tdfG, putative TonB-dependent receptor | FT | 53 | |||||
| NGO2109 | hpuB, hemoglobin-haptoglobin utilization protein B | FT | 53 | |||||
| RTX toxin/virulence | NMB0364 | FrpA/C-related protein | EM | 44 | ||||
| NMB0584 | FrpA/C-related protein | EM | 44 | |||||
| NMB1405 | FrpA/C-related protein | EM | 44 | |||||
| NMB1412 to -1414 | FrpA/C-related protein | EM | 44 | |||||
| NGO0275 | IgA1 protease | FT | 53 | |||||
| Opa (opacity-associated protein) A–K | EM | 86 | ||||||
| NGO1822 | secY | EM | 86 | |||||
| Adaption/stress response | NGO0449 | sodB | EM | 86 | ||||
| NGO0652 | Thioredoxin I | FT | 53 | |||||
| NMB0544 | dnaK, heat shock protein, chaperone | FP | 22 | |||||
| NMB1472 | clpB, chaperone | FP | 22 | |||||
| NMB0740 | recN, DNA repair protein | EM | 44 | NGO0318 | recN | EM | 86 | |
| NMB0101 | Putative transposase | EM | 44 | |||||
| NMB1798 | Putative transposase | EM | 44 | |||||
| Energy metabolism | NMB1395 and -1396 | Alcohol dehydrogenase/mutY, A/G-specific adenine glycosylase | EM, FP | 22, 44 | ||||
| NMB1377 | lldD, l-lactate dehydrogenase | EM, FP | 22, 44 | |||||
| NMB1458 | fumC, fumarase C in TCA cycle | EM, FP | 22, 44 | NGO1029 | fumC, fumarase C in TCA cycle | EM | 86 | |
| NGO0108 | Putative oxidoreductase | FT | 53 | |||||
| NGO0114 | Putative glutaredoxin | FT | 53 | |||||
| Biosynthesis | NMB1898 | mlp, lipoprotein | EM | 44 | ||||
| NMB0317 | 7-cyano-7-deazaguanine reductase | EM | 44 | |||||
| NMB0294 | dsbA-2, disulfide interchange protein | EM | 44 | |||||
| NMB0343 | YciI-like protein | EM | 44 | |||||
| NMB0394 | nadA, quinolinate synthetase | EM | 44 | |||||
| NMB0396 | nadC, nicotinate-nucleotide pyrophosphorylase | EM | 44 | |||||
| NMB1381 | HesB/YadR/YfhF family protein | EM | 44 | |||||
| NMB1380 | nifU, nitrogen fixation | EM | 44 | |||||
| Regulators | nrrF, small RNA, transcriptional regulator | EM | 65, 66 | nrrF | P | 28 | ||
| NGO0025 | mpeR, AraC-like regulator | FT | 53 | |||||
| Hypothetical | NMB0034 to -0036 | Hypothetical protein | EM, FP | 22, 44 | ||||
| NMB0744 | Hypothetical protein | EM | 44 | NGO0322 | Hypothetical protein | FT | 53 | |
| NMB0821 | Hypothetical protein | EM | 44 | |||||
| NMB0865 and -0864 | Hypothetical protein | EM | 44 | |||||
| NMB1340 | Hypothetical protein | EM | 44 | |||||
| NMB1491 | Hypothetical protein | EM | 44 | |||||
| NMB1796 | Hypothetical protein | FP | 22 | |||||
| NMB1879 and -1880 | Hypothetical protein | FP | 22 | |||||
| NGO0554 | Hypothetical protein | FT | 53 | |||||
| Direct activation | ||||||||
| Iron acquisition/storage | NGO1205 | Putative TonB-dependent receptor | EM, FP | 105 | ||||
| NGO0794 | bfrA, bacterioferritin | EM, FP, FT | 53, 105 | |||||
| Energy metabolism | NMB1613 | fumB, fumarate hydratase | FP | 22 | ||||
| NMB0242 to -0244 | nuoB-nuoD, NADH dehydrogenase subunits | EM, FP | 23, 44 | NGO1748 to -1751 | nuo operon | EM, FP, FT | 53, 105 | |
| NGO0711 | Alcohol dehydrogenase | EM, FP, FT | 53, 105 | |||||
| NGO2116 | ATP-binding protein | EM, FP, FT | 53, 105 | |||||
| NGO0076 | Putative phosphatase | EM, FP, FT | 53, 105 | |||||
| Adaption/stress response | NMB0663 | nspA, neisserial surface protein A | EM, FP | 44, 90 | NGO0233 | nspA | EM, FP | 105 |
| NMB1622 | norB, nitric oxide reductase | FP | 23 | NGO1275 | norB | EM, FP, FT | 53, 105 | |
| NMB1623 | aniA, nitrite reductase | EM, FP | 23, 44 | NGO1276 | aniA | EM, FP, FT | 53, 105 | |
| NMB0884 | sodB, superoxide dismutase | EM | 44 | |||||
| NMB0216 | kat, catalase | EM | 44 | |||||
| NMB1436 to -1438 | Hypothetical proteins | EM, FP | 22, 44 | NGO0904 to -0906 | Hypothetical Fe-S protein; hypothetical protein; Fe-S oxidoreductase | EM, FP, FT | 53, 105 | |
| NGO1317 | Transposase | EM, FP, FT | 53, 105 | |||||
| Transcription/regulation | NGO0199 | Transcription termination factor Rho | EM, FP | 105 | ||||
| NGO1851 | DNA direct RNA polymerase subunit β | EM, FP | 105 | |||||
| Hypothetical | NMB0298 | Hypothetical protein | EM | 44 | ||||
| NGO1207 to -1209 | Excinuclease ABC subunit A; restriction endonuclease R.NgoMIII; DNA cytosine methyltransferase M.NgoMIII | EM, FP | 105 | |||||
| NGO1282 | Hypothetical protein | EM, FP | 105 | |||||
| NGO1430 | Hypothetical protein | EM, FP, FT | 53, 105 | |||||
EM, electrophoresis mobility shift assay (EMSA): purified protein is incubated with radiolabeled probes (DNA or RNA) and subsequently run on a native polyacrylamide gel. Protein-bound probes show less mobility and shift up compared to free probes. FP, footprinting: DNase I is used to cut one end of labeled DNA, and the resulting patterns are analyzed by gel electrophoresis. The protein-bound site on the DNA is protected from cleavage and results in a clear area. FT (FurTa) (99), Fur titration assay: a bacterial genomic DNA library is constructed on a multicopy plasmid, such as puc18. The plasmids are transformed into an E. coli strain deficient in enterochelin synthesis and containing a fusion construct of the promoter of fhuF::lacZ in the chromosome. The promoter of fhuF has weak affinity to the FurFe2+ repressor. If the multicopy plasmids do not contain a Fur box, FurFe2+ represses the promoter region of fhuF::lacZ fusion construct so that the strain produces Lac (white) colonies on MacConkey plates supplemented with iron. In contrast, if the multicopy plasmids contain a Fur box, then the high number of these Fur boxes competes with the binding of FurFe2+ to derepress the promoter region of fhuF::lacZ fusion construct, which results in Lac-positive colonies on MacConkey plates supplemented with iron.
lbpA and lbpB genes are not present in N. gonorrhoeae strain FA1090 but were identified in strain FA19.
Predicted to be directly regulated by Fur according to the presence of an in silico Fur box in the promoter region of the gene.
Fur REGULON OF N. GONORRHOEAE
Microarray studies in N. gonorrhoeae determined that ∼20% of the gonococcal genome is regulated in response to growth under iron-replete versus -depleted conditions (27, 53). When examined by in silico analysis, 92 genes or operons were predicted to contain a Fur box (27, 53). However, only a small percentage of these putative operator regions were demonstrated to bind Fur by a Fur titration assay (FurTa) (99), EMSA, and/or footprinting (Table 2) (25, 36, 41, 53, 105). Similar to N. meningitidis, gonococcal Fur-repressed genes include a large number involved in iron acquisition (Table 2). However, only three genes or loci encoding proteins for energy metabolism and biosynthesis (fumC, NGO0108, and NGO0114) are identified as being Fur repressed in N. gonorrhoeae (Table 2) (53, 86). The Fur-repressed virulence-associated genes and stress response genes are also different from those in N. meningitidis (Table 2). Genes encoding IgA1 protease, which cleaves human IgA on the mucosal surface (88), SecY, a putative preprotein translocase (86), and the opacity-associated Opa proteins (A to K) are directly repressed by Fur only in N. gonorrhoeae (Table 2), although N. meningitidis also contains these genes (18, 90). Interestingly, sodB, which is one of the Fur-activated genes in N. meningitidis, is repressed by Fur in the gonococcus (Table 2). In addition to sodB, only one gene and one locus, recN and NGO0652, are identified as stress response genes in N. gonorrhoeae (Table 2). NGO0652 encodes a putative thioredoxin protein (53), and a thioredoxin-like protein in the gonococcus has been suggested to play a role in defense against oxidative stress (1). More Fur-activated genes and operons have been identified in the gonococci than in the meningococci to date (Table 2). In addition to nspA, norB, aniA, and the nuo operon that have already been reported in the meningococcus, Fur-activated genes and loci in the gonococcus include those involved in iron storage and transport (NGO1205 and bfrA), transcription and regulation (NGO0199 and NGO1851), energy metabolism (NGO0711 and NGO2116), and adaption (NGO0076 and NGO1317) (Table 2) (53, 105). Their roles in gonococcal pathogenesis have yet to be investigated.
MECHANISMS OF Fur-MEDIATED DIRECT AND INDIRECT REGULATION IN PATHOGENIC NEISSERIA
Direct Fur repression.
The repression mechanism of Fur, as well as the Fur box consensus sequence, is similar to that determined in other bacterial species (22, 25, 36, 43, 44, 86). Fur dimers bind to the −10 and −35 motifs in the promoter region to prevent the binding of RNA polymerase, which results in transcriptional repression of the gene. The apo-Fur repression mechanism may also be present in Neisseria, since several genes of N. gonorrhoeae have been shown to be repressed by Fur under iron-depleted conditions (105).
Direct Fur activation.
The mechanism by which Fur functions to directly activate gene transcription is poorly understood. In general, proteins which function to activate transcription utilize one of the following three pathways: (i) the activator forces a repressor out of a potential binding site, allowing the initiation of transcription; (ii) the activator recruits RNA polymerase to enhance transcription; or (iii) the activator binding alters DNA morphology, allowing RNA polymerase binding (11). In addition, multiple mechanisms may be utilized simultaneously (11). Thus, in contrast to the simple mechanism of Fur-mediated repression, Fur-mediated direct activation may result from several different pathways.
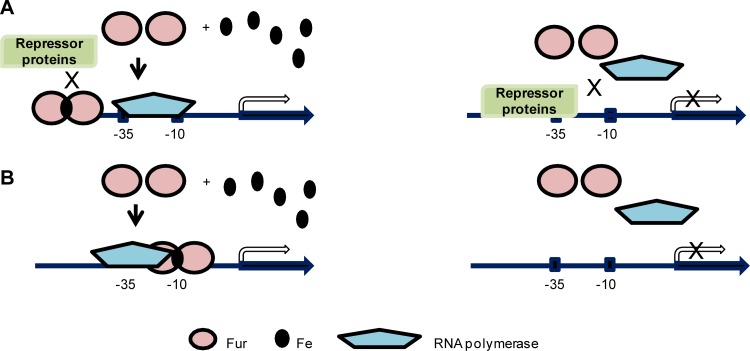
Examples of the first pathway have been characterized in Neisseria. In N. gonorrhoeae, Fur-mediated activation of the norB gene is accomplished via the Fur box overlapping with the binding site of another repressor, ArsR. In this scenario, Fur binding to the promoter region of norB competes with ArsR and results in derepression of transcription (50). The position of the Fur box in the promoter region is at a distance further upstream from the −10 and −35 motifs than those in the genes repressed by Fur (Fig. 3A). Similar positioning of Fur boxes were reported in aniA, norB, and nuoA in N. meningitidis and in NGO0199, NGO1275 (norB), NGO1276 (aniA), and NGO1282 in N. gonorrhoeae (22, 105), suggesting that these genes could be activated by Fur via exclusion of a repressor. Interestingly, recent identifications of genes directly activated by Fur and characterization of their respective Fur boxes have shown that the Fur boxes of a majority of these genes in N. gonorrhoeae are in fact localized close to the −10 and −35 motifs, either overlapping both motifs or downstream of the −10 motif (105). This suggests an alternative mechanism utilized by Fur to directly initiate transcription. It is hypothesized that binding of Fur to a position close to the −10 and −35 motifs may recruit RNA polymerase binding in order to enhance transcription initiation (Fig. 3B) (105). Depending on the subtle differences of the positions of Fur box relative to the promoter motifs, Fur may interact with different subunits of RNA polymerase, although evidence for the direct interaction between Fur and subunits of RNA polymerase has not yet been established. However, we cannot rule out the possibility that Fur may utilize unidentified mechanisms other than ones discussed above to activate gene transcription.
Fig 3.
Fur mediates direct activation via variable mechanisms. (A) An iron-bound Fur dimer binds to a site further upstream of −10 and −35 motifs in the promoter region and out-competes binding of a repressor protein, thus activating transcription (left panel). Without iron, Fur does not function and the repressor protein binds to the promoter region to repress gene transcription (right panel). Three examples have been reported in E. coli and N. gonorrhoeae (50, 77). (B) An iron-bound Fur dimer binds to a site overlapping the −10 and −35 motifs in the promoter region in order to recruit RNA polymerase to activate transcription (left panel). Without iron, Fur does not bind to the promoter region and RNA polymerase is not recruited to initiate transcription (right panel). This mechanism is still a hypothesized model without experimental evidence (105).
Indirect Fur regulation.
Fur-mediated indirect regulation in pathogenic Neisseria can occur via sRNA-mediated regulation or a regulator protein. Fur-mediated activation has been demonstrated to function indirectly through repressing a sRNA, NrrF, in N. meningitidis, which negatively regulates its target genes (65, 66). Nearly all sRNAs described in a variety of Gram-positive and Gram-negative bacteria share four common characteristics: (i) they are mainly localized in intergenic regions; (ii) the sequences are highly conserved among most genetically similar species; (iii) the 3′ termini of sRNAs usually contain a Rho-independent terminator structure, which is composed of a stem-loop followed by a polyuridine region; and (iv) the lengths of sRNAs range from 50 to 300 nucleotides (100). Generally, sRNAs base pair to the ribosome-binding site on target mRNAs to interfere with ribosome binding and block the initiation of translation (100). The complementarity between a sRNA and mRNA requires a “seed” region of at least 8 to 9 continuous base pairs (100). Alternatively, sRNAs may lead to decreased stability of target mRNAs, resulting in reduced translation (100). In addition, many of the known sRNAs require a protein cofactor, Host Factor Qβ-phage (Hfq), to facilitate the binding of sRNAs to mRNA (42, 97).
The meningococcal sRNA NrrF (between loci NMB2073 and NMB2074) was identified by screening intergenic regions for Fur boxes upstream of Rho-independent terminators (65). One of 19 possible candidates, NrrF, was found to be iron regulated via Fur (65). NrrF is upregulated under iron-depleted conditions in a wild-type strain and derepressed in the fur mutant strain in a manner independent of the presence of iron, as determined by both Northern blot and reverse transcription-PCR (RT-PCR) experiments (65). The regulatory targets of NrrF were predicted using a bioinformatics approach which identified the sdhA-sdhC operon as a possible target (65). Subsequent experiments examining transcription of sdhA-sdhC in a wild-type strain and a nrrF mutant strain under iron-depleted or iron-replete conditions confirmed an NrrF-dependent repression pattern for sdhA-sdhC (65). Interestingly, unlike the Fur-regulated sRNAs in other bacteria, NrrF can function in the absence of Hfq. In an Δhfq strain, the regulation of sdhA-sdhC in response to iron availability is unchanged, as is the stability of NrrF (66). NrrF is the first Fur-controlled negative regulator that has been discovered in pathogenic Neisseria (65, 66). Targets of NrrF, sdhA-sdhC, are in turn indirectly activated by Fur (65, 66). A NrrF homologue (between NGO2002 and NGO2004) has also been found in the N. gonorrhoeae genome (28).
To date, one Neisseria regulatory protein which is under Fur-mediated direct regulation has been reported. The N. gonorrhoeae Fur-repressed MpeR protein, an AraC-like regulator (53), activates fetA, an outer membrane transporter required for acquisition of xenosiderophore ferric enterobactin as an iron source (47) and represses MtrF and MtrR, which function in the mtr efflux pump and modulate antimicrobial resistance systems (34, 67, 89).
Fur GLOBAL REGULATORY CIRCUITS AND CROSS-TALK WITH NEWLY DEFINED REGULONS
With the discovery of increasing numbers of Fur-regulated genes, additional regulators have been found to cooperate with Fur to control the same gene or operon. For example, in N. meningitidis, Fur-regulated hemO and hmbR were shown to be positively regulated by the two-component system MisR/S under both iron-depleted and -replete conditions (106). Transcription of N. meningitidis kat, which is activated by Fur, is also repressed by OxyR in the absence of H2O2 and activated by OxyR with H2O2 (49). Perhaps the best-studied cooperative regulation has been shown for the gonococcal aniA and norB genes, involved in the anaerobic respiration pathway in N. gonorrhoeae. Several regulators are utilized for aniA and norB regulation, including Fur, NsrR, NarQ/P, FNR, and ArsR (50, 52). NsrR is a repressor containing a [2Fe-2S] cluster, which is involved in NO sensing (50, 52). In addition to iron limitation, the pathogenic Neisseria spp. may also encounter other stresses in the host environment, such as the simultaneous presence of NO, H2O2, and pH. The gonococcal Fur regulon has also been shown to overlap anaerobic and hydrogen peroxide regulons (51, 92). Thus, it is highly likely that Fur functions together with other regulators to enable Neisseria spp. to respond to complicated environmental conditions or stimuli within the human host.
We propose that the Neisseria Fur regulon encompasses a complicated network due to the ability of this protein to function as either a repressor or an activator in both direct and indirect pathways. To date, studies in the pathogenic Neisseria have examined only a small subset of Fur-regulated genes and have had limited value in deciphering a Fur-mediated global network. High-throughput bioinformatics techniques designed to assist in the analysis of the relationships among regulons of different regulators (19) should help to identify the entire Neisseria Fur regulon.
BIOLOGICAL ROLES OF Fur REGULATORY CIRCUITS
Although the genomes of N. meningitidis and N. gonorrhoeae are closely related, the Fur regulons of these two organisms are not completely overlapping (Table 2). These differences may relate to pathogen-specific requirements during human colonization and associated inflammatory pathologies.
N. gonorrhoeae mainly colonizes the human urethra, endocervix, fallopian tubes, and uterus. This pathogen causes urethritis in men, with obvious inflammatory symptoms such as a purulent discharge with an influx of polymorphonuclear leukocytes (PMN). In women, gonococcal infection presents as cervicitis, vaginitis, or a more serious pelvic inflammatory disease. Also, infection in women is typically asymptomatic and may lead to serious complications, including endometritis, salpingitis, and disseminated gonococcal infection (DGI) (30). N. meningitidis frequently colonizes the nasopharynx and causes meningitis or septicemia upon entering the cerebrospinal fluid or bloodstream, respectively. Generally, host niches for both organisms are iron-depleted environments, as free iron in the human host is scarce. Therefore, genes involved in iron acquisition, including fur itself, are upregulated in both pathogens during N. meningitidis infection in human blood and in the N. gonorrhoeae RNA isolated from cervical swab specimens from women with uncomplicated gonorrhea or urethral swab specimens from men with urethral infections (2, 3, 29). However, one of the differences between species-specific host niches is the iron source. In serum, transferrin is the major iron-carrying protein, while in the mucosal surfaces, lactoferrin is the major iron source (14). In addition, the concentration of lactoferrin can change with the menstrual cycle in human vaginal mucus (15). All N. meningitidis strains are able to utilize both transferrin and lactoferrin (70, 71), which may guarantee the survival of N. meningitidis in both types of host niches. In contrast, not all N. gonorrhoeae strains contain lbpA and lbpB genes, required for utilizing lactoferrin (8, 9, 41, 70). It has been proposed that gonococcal strains unable to utilize lactoferrin may be related to the asymptomatic infections often observed in women (10).
In N. meningitidis, the aniA (nitrite reductase), kat (catalase), and nspA (Neisseria surface protein A) genes are under direct Fur activation and have been demonstrated to be upregulated during N. meningitidis colonization in human blood (29). The kat and aniA genes have been postulated to play a role in N. meningitidis survival under conditions of stress from reactive oxygen and nitrogen species from neutrophils and macrophages (4, 87). Interestingly, the kat gene is not under iron or Fur-mediated control in the gonococcus (53). The nspA gene encodes a human factor H binding protein that facilitates resistance to human complement, resulting in enhanced survival of N. meningitidis in blood (29, 60). Furthermore, genes under Fur-mediated direct activation, including nspA and aniA, were not detected in specimens isolated from women with gonococcal infection (3). Another significant difference between N. meningitidis and N. gonorrhoeae Fur regulons during infection may relate to the expression of Opa proteins, which are involved in bacterial adherence and invasion of human epithelial cells and neutrophils (18). N. meningitidis contains 3 to 4 Opa proteins, and at least one of them (NMB1636) is upregulated in human blood (29), although none of meningococcal opa genes are regulated via Fur (22, 44). In contrast, the putative promoter regions of all 11 gonococcal opa genes (A to K) have been shown to bind directly to Fur (86). Gonococcal Opa proteins have been shown to be expressed during natural infections as well as in experimentally infected volunteers (54, 55, 93). Furthermore, Opa–carcinoembryonic antigen-related cell adhesion molecule (CEACAM) interactions have been shown to promote gonococcal colonization in mouse models (16, 75, 76, 82). Above all, these results suggest that specific colonization niches and the associated pathogenic processes of the two pathogenic species serve to define the Fur regulons of these organisms and, in particular, those genes which are activated by Fur.
Moreover, the global effects resulting from Fur-mediated regulation determine the virulence of several bacterial pathogens. A large number of genes encoding iron-uptake protein homologues in the Staphylococcus aureus genome contain a predicted Fur box in their promoter regions, suggesting a major role of Fur in iron homeostasis (48). A fur mutant strain of S. aureus showed growth defects and higher sensitivity to H2O2 (48). These characteristics may have led to the reduced virulence (lower recovery of the fur mutant strain compared to the wild-type strain) seen in a murine skin abscess model (48). Fur of Bacillus cereus, an opportunistic human pathogen that causes food poisoning and endophthalmitis, may control at least 16 genes, according to the results of Fur-box prediction in the promoter regions of the genome (46). These genes include those involved in iron uptake and storage, secondary cellular metabolism, and virulence. The fur mutant strain of B. cereus also shows reduced virulence, with a 50% lethal dose (LD50) value of 4,932 CFU compared to an LD50 value of 1,859 CFU of the wild-type strain in an insect model (46). Similarly, V. cholerae has been shown to have at least 65 Fur iron-repressed genes which are involved in iron acquisition and metabolism and two genes indirectly activated by Fur (69). In an infant mouse model of intestinal colonization, the V. cholerae fur mutant strain displayed significantly attenuated colonization when competing with the wild-type strain (69). Fur of the gastric pathogen H. pylori regulates genes critical for acid acclimation and oxidative stress (39, 80). The fur mutant strain of H. pylori displays a 100-fold-higher 50% infectious dose than the wild-type strain and lower colonization ability when competing with the wild-type strain in the Mongolian gerbil model (39, 73, 80). In addition, the fur mutant strain showed an attenuated ability to induce host inflammation and injury (73). All of the studies noted above emphasized the importance of the Fur regulon in bacterial pathogenesis.
CONCLUDING REMARKS
Recent studies have begun to define the Fur regulons of the pathogenic Neisseria. For both N. meningitidis and N. gonorrhoeae, global mechanisms of transcriptional control by Fur have been linked to the ability of these pathogens to cause disease and to respond to various stimuli within the human host. We predict that Fur-regulated circuits embrace broad components in the genomes and enable these organisms to respond to a variety of stress situations. Thus, understanding pathogenic aspects of Fur-mediated regulation is critical in revealing bacterial pathogenic mechanisms and will help to discover new therapeutic targets in these pathogens.
Footnotes
Published ahead of print 10 August 2012
REFERENCES
- 1. Achard ME, et al. 2009. A periplasmic thioredoxin-like protein plays a role in defense against oxidative stress in Neisseria gonorrhoeae. Infect. Immun. 77:4934–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agarwal S, et al. 2005. The gonococcal Fur-regulated tbpA and tbpB genes are expressed during natural mucosal gonococcal infection. Infect. Immun. 73:4281–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Agarwal S, Sebastian S, Szmigielski B, Rice PA, Genco CA. 2008. Expression of the gonococcal global regulatory protein Fur and genes encompassing the Fur and iron regulon during in vitro and in vivo infection in women. J. Bacteriol. 190:3129–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anjum MF, Stevanin TM, Read RC, Moir JW. 2002. Nitric oxide metabolism in Neisseria meningitidis. J. Bacteriol. 184:2987–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Argaman L, et al. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941–950 [DOI] [PubMed] [Google Scholar]
- 6. Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477 [DOI] [PubMed] [Google Scholar]
- 7. Baichoo N, Helmann JD. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Biswas GD, Anderson JE, Chen CJ, Cornelissen CN, Sparling PF. 1999. Identification and functional characterization of the Neisseria gonorrhoeae lbpB gene product. Infect. Immun. 67:455–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Biswas GD, Sparling PF. 1995. Characterization of lbpA, the structural gene for a lactoferrin receptor in Neisseria gonorrhoeae. Infect. Immun. 63:2958–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Britigan BE, Cohen MS, Sparling PF. 1985. Gonococcal infection: a model of molecular pathogenesis. N. Engl. J. Med. 312:1683–1694 [DOI] [PubMed] [Google Scholar]
- 11. Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57–65 [DOI] [PubMed] [Google Scholar]
- 12. Butcher BG, et al. 2011. Characterization of the Fur Regulon in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 193:4598–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carpenter BM, Whitmire JM, Merrell DS. 2009. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect. Immun. 77:2590–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen CY, Genco CA, Rock JP, Morse SA. 1989. Physiology and metabolism of Neisseria gonorrhoeae and Neisseria meningitidis: implications for pathogenesis. Clin. Microbiol. Rev. 2(Suppl):S35–S40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen MS, Britigan BE, French M, Bean K. 1987. Preliminary observations on lactoferrin secretion in human vaginal mucus: variation during the menstrual cycle, evidence of hormonal regulation, and implications for infection with Neisseria gonorrhoeae. Am. J. Obstet. Gynecol. 157:1122–1125 [DOI] [PubMed] [Google Scholar]
- 16. Cole JG, Fulcher NB, Jerse AE. 2010. Opacity proteins increase Neisseria gonorrhoeae fitness in the female genital tract due to a factor under ovarian control. Infect. Immun. 78:1629–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Craig SA, Carpenter CD, Mey AR, Wyckoff EE, Payne SM. 2011. Positive regulation of the Vibrio cholerae porin OmpT by iron and fur. J. Bacteriol. 193:6505–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Criss AK, Seifert HS. 2012. A bacterial siren song: intimate interactions between Neisseria and neutrophils. Nat. Rev. Microbiol. 10:178–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Danielli A, Scarlato V. 2010. Regulatory circuits in Helicobacter pylori: network motifs and regulators involved in metal-dependent responses. FEMS Microbiol. Rev. 34:738–752 [DOI] [PubMed] [Google Scholar]
- 20. Davies BW, Bogard RW, Mekalanos JJ. 2011. Mapping the regulon of Vibrio cholerae ferric uptake regulator expands its known network of gene regulation. Proc. Natl. Acad. Sci. U. S. A. 108:12467–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. 2005. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 187:4005–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delany I, Grifantini R, Bartolini E, Rappuoli R, Scarlato V. 2006. Effect of Neisseria meningitidis fur mutations on global control of gene transcription. J. Bacteriol. 188:2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delany I, Rappuoli R, Scarlato V. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52:1081–1090 [DOI] [PubMed] [Google Scholar]
- 24. de Lorenzo V, Wee S, Herrero M, Neilands JB. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Desai PJ, Angerer A, Genco CA. 1996. Analysis of Fur binding to operator sequences within the Neisseria gonorrhoeae fbpA promoter. J. Bacteriol. 178:5020–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dian C, et al. 2011. The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol. Microbiol. 79:1260–1275 [DOI] [PubMed] [Google Scholar]
- 27. Ducey TF, Carson MB, Orvis J, Stintzi AP, Dyer DW. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae by microarray analysis in defined medium. J. Bacteriol. 187:4865–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ducey TF, Jackson L, Orvis J, Dyer DW. 2009. Transcript analysis of nrrF, a Fur repressed sRNA of Neisseria gonorrhoeae. Microb. Pathog. 46:166–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Echenique-Rivera H, et al. 2011. Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathog. 7:e1002027 doi:10.1371/journal.ppat.1002027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edwards JL, Apicella MA. 2004. The molecular mechanisms used by Neisseria gonorrhoeae to initiate infection differ between men and women. Clin. Microbiol. Rev. 17:965–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ernst JF, Bennett RL, Rothfield LI. 1978. Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J. Bacteriol. 135:928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Escolar L, de Lorenzo V, Perez-Martin J. 1997. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol. Microbiol. 26:799–808 [DOI] [PubMed] [Google Scholar]
- 33. Escolar L, Perez-Martin J, de Lorenzo V. 1998. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283:537–547 [DOI] [PubMed] [Google Scholar]
- 34. Folster JP, Shafer WM. 2005. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J. Bacteriol. 187:3713–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Forman S, et al. 2003. Neisseria meningitidis RTX proteins are not required for virulence in infant rats. Infect. Immun. 71:2253–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forng RY, Ekechukwu CR, Subbarao S, Morse SA, Genco CA. 1997. Promoter mapping and transcriptional regulation of the iron-regulated Neisseria gonorrhoeae fbpA gene. J. Bacteriol. 179:3047–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fuangthong M, Helmann JD. 2003. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 185:6348–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gaballa A, et al. 2008. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. U. S. A. 105:11927–11932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gancz H, Censini S, Merrell DS. 2006. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74:602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao H, et al. 2008. The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 190:3063–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Genco CA, Desai PJ. 1996. Iron acquisition in the pathogenic Neisseria. Trends Microbiol. 4:179–184 [DOI] [PubMed] [Google Scholar]
- 42. Gottesman S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol. 58:303–328 [DOI] [PubMed] [Google Scholar]
- 43. Grifantini R, et al. 2004. Characterization of a novel Neisseria meningitidis Fur and iron-regulated operon required for protection from oxidative stress: utility of DNA microarray in the assignment of the biological role of hypothetical genes. Mol. Microbiol. 54:962–979 [DOI] [PubMed] [Google Scholar]
- 44. Grifantini R, et al. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. U. S. A. 100:9542–9547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hantke K. 1984. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol. Gen. Genet. 197:337–341 [DOI] [PubMed] [Google Scholar]
- 46. Harvie DR, Vilchez S, Steggles JR, Ellar DJ. 2005. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology 151:569–577 [DOI] [PubMed] [Google Scholar]
- 47. Hollander A, Mercante AD, Shafer WM, Cornelissen CN. 2011. The iron-repressed, AraC-like regulator MpeR activates expression of fetA in Neisseria gonorrhoeae. Infect. Immun. 79:4764–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Horsburgh MJ, Ingham E, Foster SJ. 2001. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ieva R, et al. 2008. OxyR tightly regulates catalase expression in Neisseria meningitidis through both repression and activation mechanisms. Mol. Microbiol. 70:1152–1165 [DOI] [PubMed] [Google Scholar]
- 50. Isabella V, et al. 2008. cis- and trans-acting elements involved in regulation of norB (norZ), the gene encoding nitric oxide reductase in Neisseria gonorrhoeae. Microbiology 154:226–239 [DOI] [PubMed] [Google Scholar]
- 51. Isabella VM, Clark VL. 2011. Deep sequencing-based analysis of the anaerobic stimulon in Neisseria gonorrhoeae. BMC Genomics 12:51 doi:10.1186/1471-2164-12-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Isabella VM, Lapek JD, Jr, Kennedy EM, Clark VL. 2009. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol. Microbiol. 71:227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jackson LA, et al. 2010. Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon. J. Bacteriol. 192:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. James JF, Swanson J. 1978. Studies on gonococcus infection. XIII. Occurrence of color/opacity colonial variants in clinical cultures. Infect. Immun. 19:332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jerse AE, et al. 1994. Multiple gonococcal opacity proteins are expressed during experimental urethral infection in the male. J. Exp. Med. 179:911–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ledala N, Sengupta M, Muthaiyan A, Wilkinson BJ, Jayaswal RK. 2010. Transcriptomic response of Listeria monocytogenes to iron limitation and Fur mutation. Appl. Environ. Microbiol. 76:406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee HJ, Bang SH, Lee KH, Park SJ. 2007. Positive regulation of fur gene expression via direct interaction of fur in a pathogenic bacterium, Vibrio vulnificus. J. Bacteriol. 189:2629–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499 [DOI] [PubMed] [Google Scholar]
- 59. Lewin AC, Doughty PA, Flegg L, Moore GR, Spiro S. 2002. The ferric uptake regulator of Pseudomonas aeruginosa has no essential cysteine residues and does not contain a structural zinc ion. Microbiology 148:2449–2456 [DOI] [PubMed] [Google Scholar]
- 60. Lewis LA, et al. 2010. The meningococcal vaccine candidate neisserial surface protein A (NspA) binds to factor H and enhances meningococcal resistance to complement. PLoS Pathog. 6:e1001027 doi:10.1371/journal.ppat.1001027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Massé E, Escorcia FE, Gottesman S. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Massé E, Majdalani N, Gottesman S. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6:120–124 [DOI] [PubMed] [Google Scholar]
- 64. Massé E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187:6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. 2007. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 189:3686–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mellin JR, et al. 2010. Role of Hfq in iron-dependent and -independent gene regulation in Neisseria meningitidis. Microbiology 156:2316–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mercante AD, et al. 2012. MpeR regulates the mtr efflux locus in Neisseria gonorrhoeae and modulates antimicrobial resistance by an iron-responsive mechanism. Antimicrob. Agents Chemother. 56:1491–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Merrell DS, et al. 2003. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect. Immun. 71:6510–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. 2005. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 73:8167–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mickelsen PA, Blackman E, Sparling PF. 1982. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from lactoferrin. Infect. Immun. 35:915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mickelsen PA, Sparling PF. 1981. Ability of Neisseria gonorrhoeae, Neisseria meningitidis, and commensal Neisseria species to obtain iron from transferrin and iron compounds. Infect. Immun. 33:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Miles S, Carpenter BM, Gancz H, Merrell DS. 2010. Helicobacter pylori apo-Fur regulation appears unconserved across species. J. Microbiol. 48:378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Miles S, et al. 2010. Detailed in vivo analysis of the role of Helicobacter pylori Fur in colonization and disease. Infect. Immun. 78:3073–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mills SA, Marletta MA. 2005. Metal binding characteristics and role of iron oxidation in the ferric uptake regulator from Escherichia coli. Biochemistry 44:13553–13559 [DOI] [PubMed] [Google Scholar]
- 75. Muenzner P, Bachmann V, Zimmermann W, Hentschel J, Hauck CR. 2010. Human-restricted bacterial pathogens block shedding of epithelial cells by stimulating integrin activation. Science 329:1197–1201 [DOI] [PubMed] [Google Scholar]
- 76. Muenzner P, Rohde M, Kneitz S, Hauck CR. 2005. CEACAM engagement by human pathogens enhances cell adhesion and counteracts bacteria-induced detachment of epithelial cells. J. Cell Biol. 170:825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nandal A, et al. 2010. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe(2+)-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol. Microbiol. 75:637–657 [DOI] [PubMed] [Google Scholar]
- 78. Ochsner UA, Vasil ML. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. U. S. A. 93:4409–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Osicka R, Kalmusova J, Krizova P, Sebo P. 2001. Neisseria meningitidis RTX protein FrpC induces high levels of serum antibodies during invasive disease: polymorphism of frpC alleles and purification of recombinant FrpC. Infect. Immun. 69:5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pich OQ, Carpenter BM, Gilbreath JJ, Merrell DS. 2012. Detailed analysis of Helicobacter pylori Fur-regulated promoters reveals a Fur box core sequence and novel Fur-regulated genes. Mol. Microbiol. 84:921–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pohl E, et al. 2003. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47:903–915 [DOI] [PubMed] [Google Scholar]
- 82. Sadarangani M, Pollard AJ, Gray-Owen SD. 2011. Opa proteins and CEACAMs: pathways of immune engagement for pathogenic Neisseria. FEMS Microbiol. Rev. 35:498–514 [DOI] [PubMed] [Google Scholar]
- 83. Saito T, Duly D, Williams RJ. 1991. The histidines of the iron-uptake regulation protein, Fur. Eur. J. Biochem. 197:39–42 [DOI] [PubMed] [Google Scholar]
- 84. Schäffer S, Hantke K, Braun V. 1985. Nucleotide sequence of the iron regulatory gene fur. Mol. Gen. Genet. 200:110–113 [DOI] [PubMed] [Google Scholar]
- 85. Schielke S, Frosch M, Kurzai O. 2010. Virulence determinants involved in differential host niche adaptation of Neisseria meningitidis and Neisseria gonorrhoeae. Med. Microbiol. Immunol. 199:185–196 [DOI] [PubMed] [Google Scholar]
- 86. Sebastian S, Agarwal S, Murphy JR, Genco CA. 2002. The gonococcal fur regulon: identification of additional genes involved in major catabolic, recombination, and secretory pathways. J. Bacteriol. 184:3965–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Seib KL, Tseng HJ, McEwan AG, Apicella MA, Jennings MP. 2004. Defenses against oxidative stress in Neisseria gonorrhoeae and Neisseria meningitidis: distinctive systems for different lifestyles. J. Infect. Dis. 190:136–147 [DOI] [PubMed] [Google Scholar]
- 88. Senior BW, Stewart WW, Galloway C, Kerr MA. 2001. Cleavage of the hormone human chorionic gonadotropin, by the Type 1 IgA1 protease of Neisseria gonorrhoeae, and its implications. J. Infect. Dis. 184:922–925 [DOI] [PubMed] [Google Scholar]
- 89. Shafer WM, et al. 2001. Genetic organization and regulation of antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitidis. J. Mol. Microbiol. Biotechnol. 3:219–224 [PubMed] [Google Scholar]
- 90. Shaik YB, et al. 2007. Expression of the iron-activated nspA and secY genes in Neisseria meningitidis group B by Fur-dependent and -independent mechanisms. J. Bacteriol. 189:663–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sheikh MA, Taylor GL. 2009. Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol. Microbiol. 72:1208–1220 [DOI] [PubMed] [Google Scholar]
- 92. Stohl EA, Criss AK, Seifert HS. 2005. The transcriptome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol. Microbiol. 58:520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Swanson J, Barrera O, Sola J, Boslego J. 1988. Expression of outer membrane protein II by gonococci in experimental gonorrhea. J. Exp. Med. 168:2121–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Troxell B, Fink RC, Porwollik S, McClelland M, Hassan HM. 2011. The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol. 11:236 doi:10.1186/1471-2180-11-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Troxell B, et al. 2011. Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193:497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Tsolis RM, Baumler AJ, Stojiljkovic I, Heffron F. 1995. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J. Bacteriol. 177:4628–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Urban JH, Vogel J. 2007. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35:1018–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. van Vliet AH, et al. 2003. Differential regulation of amidase- and formamidase-mediated ammonia production by the Helicobacter pylori fur repressor. J. Biol. Chem. 278:9052–9057 [DOI] [PubMed] [Google Scholar]
- 99. Vassinova N, Kozyrev D. 2000. A method for direct cloning of fur-regulated genes: identification of seven new fur-regulated loci in Escherichia coli. Microbiology 146(Pt 12):3171–3182 [DOI] [PubMed] [Google Scholar]
- 100. Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Whitby PW, Seale TW, VanWagoner TM, Morton DJ, Stull TL. 2009. The iron/heme regulated genes of Haemophilus influenzae: comparative transcriptional profiling as a tool to define the species core modulon. BMC Genomics 10:6 doi:10.1186/1471-2164-10-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Whitby PW, Vanwagoner TM, Seale TW, Morton DJ, Stull TL. 2006. Transcriptional profile of Haemophilus influenzae: effects of iron and heme. J. Bacteriol. 188:5640–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wilderman PJ, et al. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101:9792–9797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Xiong A, Singh VK, Cabrera G, Jayaswal RK. 2000. Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus. Microbiology 146(Pt 3):659–668 [DOI] [PubMed] [Google Scholar]
- 105. Yu C, Genco CA. 2012. Fur-mediated activation of gene transcription in the human pathogen Neisseria gonorrhoeae. J. Bacteriol. 194:1730–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhao S, Montanez GE, Kumar P, Sannigrahi S, Tzeng YL. 2010. Regulatory role of the MisR/S two-component system in hemoglobin utilization in Neisseria meningitidis. Infect. Immun. 78:1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhou D, et al. 2006. Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol. Lett. 258:9–17 [DOI] [PubMed] [Google Scholar]