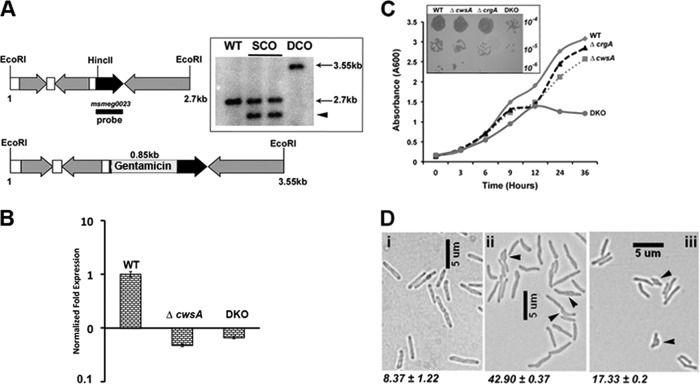
Fig 3.
Loss of CwsA affects growth and cell shape. (A) Cartoon of 2. 7-kb cwsA (msmeg0023) region of M. smegmatis (top) with an inserted copy of a gentamicin resistance cassette (bottom) (Table 1, pPP105). Black arrow, cwsA-coding region; gray arrows, coding regions of neighboring genes; white bars, intergenic regions; black bar, probe region. Genomic DNA was isolated from WT, two single-crossover colonies (lanes SCO), and one double-crossover colony (DCO), digested with EcoRI, separated by agarose gel electrophoresis, and transferred to a nitrocellulose membrane. The blot was probed with a 32P-labeled MSMEG_0023 fragment probe. The positions of expected bands in WT (2.7 kb) and double-crossover (3.55 kb) lanes are marked. The single-crossover colonies gave a band corresponding to the WT copy (2.7 kb) and another band of ∼2.3 kb (arrowhead) due to insertion of the recombination vector pPP106. (B) qRT-PCR for mutant strains. RNA was isolated from M. smegmatis WT, ΔcwsA, and DKO strains as described in the text. Quantitative real-time PCR was performed for cwsA expression levels, and data were normalized to those for housekeeping gene sigA expression (see Materials and Methods). cwsA expression levels in the two mutant strains are presented with respect to that for WT (for which the level is equal to 1). Mean expression levels ± standard deviations from two independent experiments done in triplicate are shown. (C) Growth curve for cwsA and crgA strains. Growth of M. smegmatis mC2155 (WT), ΔcrgA, ΔcwsA, and DKO strains was monitored for the indicated time points. Briefly, overnight cultures of various strains were diluted to an A600 of 0.1 and grown in 7H9 broth with albumin-dextrose, the A600 was recorded at the indicated time points, and the data were plotted. The experiment was carried out 3 times, and one representative data set is shown. (Inset) Serial dilutions of exponential cultures of indicated strains were prepared, and 5 μl was spotted on 7H10 agar medium plates and grown at 37°C. (D) Morphology of M. smegmatis and various strains. Exponential cultures of WT (i), ΔcwsA (ii), and ΔcwsA Pami::cwsAsol (iii) strains of M. smegmatis were grown in 7H9 broth with albumin-dextrose and imaged by bright-field microscopy. Arrowheads, bulgy cells. Numbers below each panel indicate percent average number of bulgy cells ± standard error from two independent experiments. Each experiment included ≥250 cells from each of the three strains.