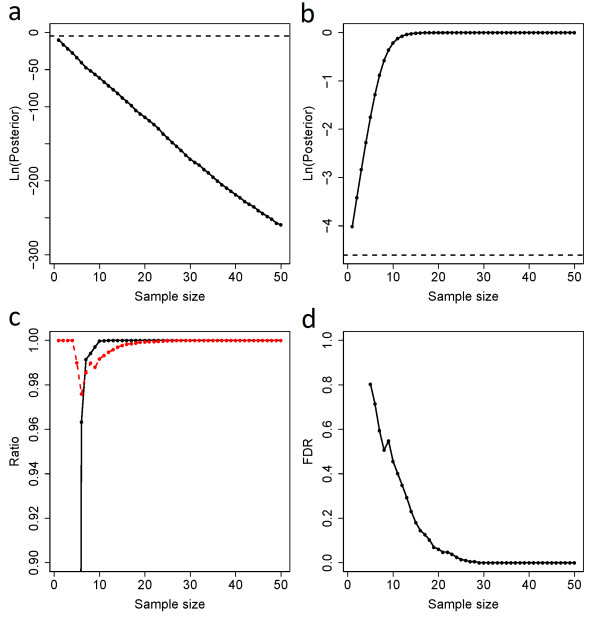
Figure 1.
Simulation-based performance analysis of dsPIG.a, b, Simulated (natural log-transformed) posteriors of (a) biallelically expressed genes and (b) imprinted genes. The dashed line in both panels stands for the log-transformed prior (0.01). Results in (a) and (b) were based on 20,000-time simulations with geometric mean as the method of averaging posteriors. c, Sensitivity (the black solid line) and specificity (the read dashed line) of our model. d, the FDR of our predictions. When sample size was <5, the FDR was not computable as sensitivity and specificity were both 0. Results in (c) and (d) were based on 20,000-time simulations with geometric mean as the method of averaging posteriors.