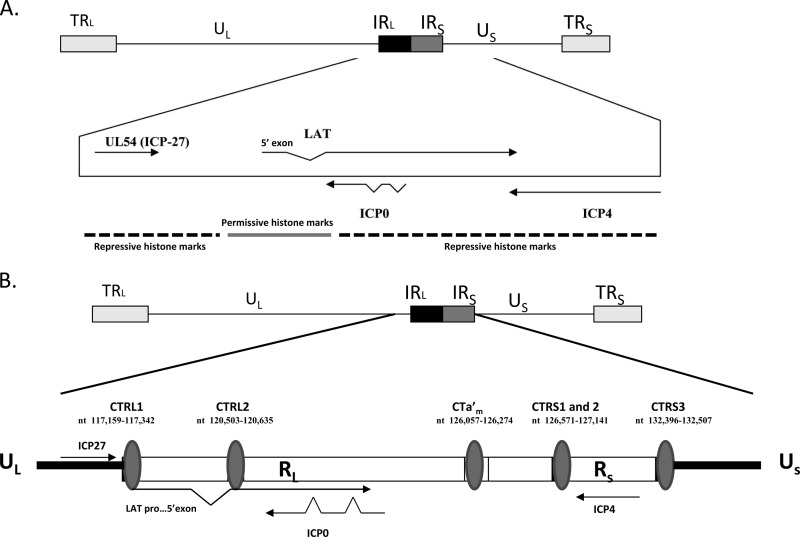
Fig 1.
The latent HSV-1 genome. (A) Regions of repressed (heterochromatin) and permissive (euchromatin) chromatin juxtaposed within the HSV-1 genome. TRL, terminal repeat long; UL, unique long region; US, unique short region; IRL, inverted long repeat; IRS, inverted short repeat; TRS, terminal repeat short. (B) Previously identified regions of the HSV-1 genome containing CCCTC and CTCCC repeats for CTCF binding flanking the LAT reactivation-critical region (CTRL1 and CTRL2), ICP0 (CTa′m), and ICP4 (CTRS1/2 and CTRS3) (2). Note that CTRS1/2 is actually 2 CTCF binding clusters separated by less than 100 nucleotides. An additional CTCF motif (CTUS1) was identified by Amelio et al. (2) (not shown).