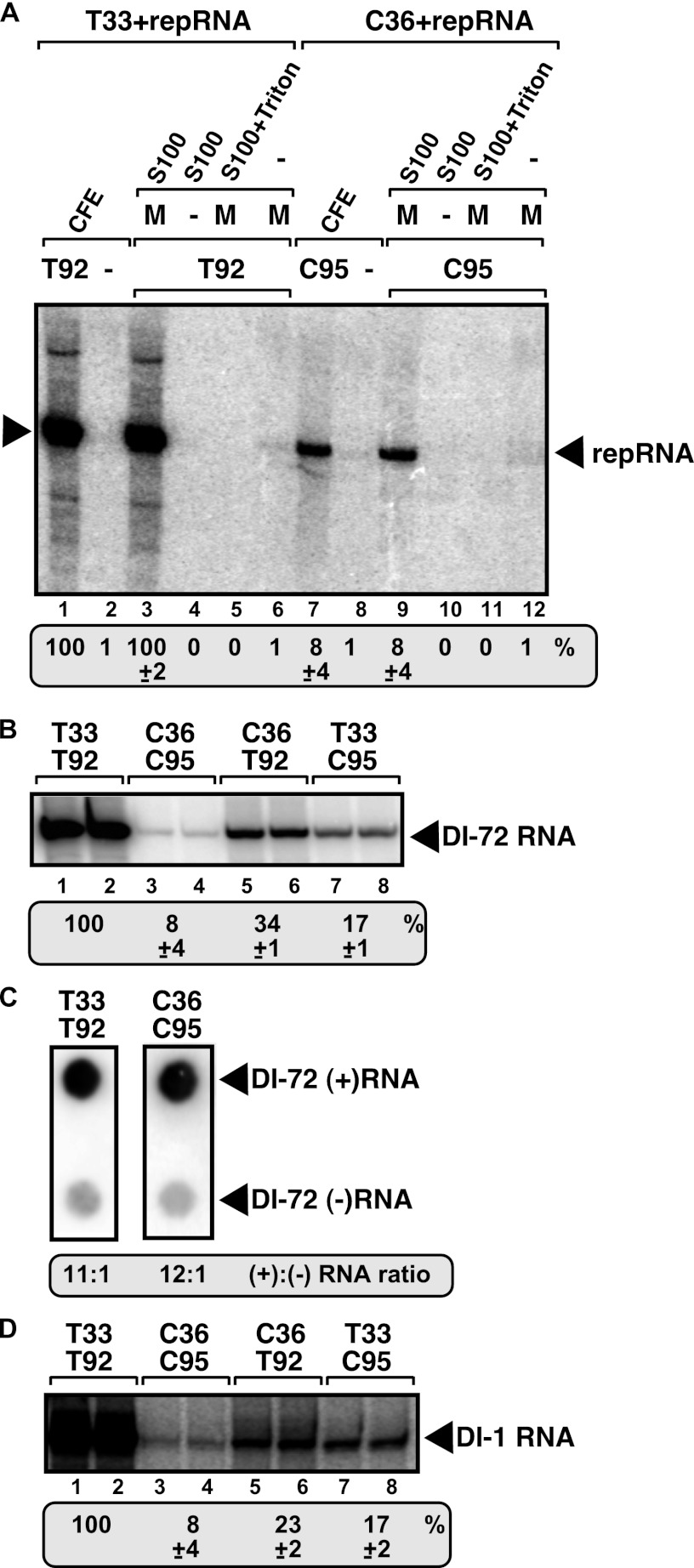
Fig 1.
In vitro reconstitution of the CIRV replicase in yeast cell extract. (A) Purified recombinant p33 (named T33) and p92pol (named T92) replication proteins of TBSV or purified recombinant p36 (named C36) and p95pol (named C95) replication proteins of CIRV in combination with the TBSV-derived DI-72 (+)repRNA were added to the cell extract (lanes 1 and 7), to the membrane plus soluble fractions (lanes 3 and 9), to the soluble fraction (lanes 4 and 10), to the 1% Triton-treated membrane plus soluble fractions (lanes 5 and 11), and to the membrane fraction (lanes 6 and 12) of the yeast cell extract. The CFE-based replication assay mixture lacked T92 (lane 2) or C95 (lane 8) as a negative control. Denaturing PAGE analysis of the 32P-labeled repRNA products obtained is shown. The full-length repRNA is indicated by an arrowhead. The result of the CFE-based replication assay with T33 and T92 was chosen as 100% (lane 1). (B) The heterologous combinations of TBSV and CIRV replication proteins are functional in the CFE-based replication assay. The activity of the reconstituted tombusvirus replicases is estimated as for panel A. Denaturing PAGE analysis of the replicase products is as shown in panel A. (C) Detection of plus- and minus-stranded RNA products produced by the reconstituted TBSV and CIRV replicases in the CFE-based replication assay. The blot contains the same amounts of cold plus- and minus-strand DI-72 RNA, while the 32P-labeled repRNA probes were generated in the CFE-based replication assay. Note that we used 5 times more CIRV replication products than TBSV to increase the signal. The ratio of plus- and minus-strand RNA products was estimated. (D) The heterologous combinations of TBSV and CIRV replication proteins with the CIRV-derived DI-1 repRNA are functional in the CFE-based replication assay. See further details described for panel B. Each experiment was repeated.