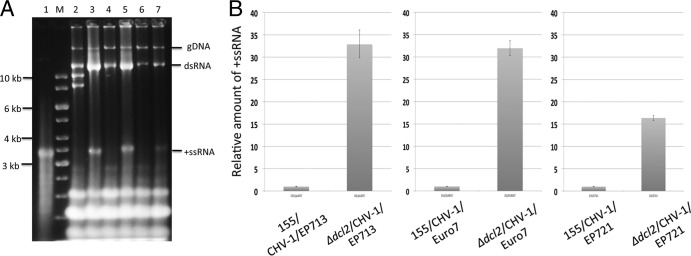
Fig 3.
Hypovirus RNA accumulation in the C. parasitica Δdcl2 mutant strain. (A). Gel analysis of RNA isolated from hypovirus-infected wild-type C. parasitica strain EP155 and the Δdcl2 mutant strain. Approximately 10 μg of total nucleic acids isolated from each infected strain was loaded and subjected to electrophoresis in 1% agarose gels set at 15 V overnight. Lane 1 contains 0.5 μg of the in vitro-synthesized 12,734-nt transcript from infectious CHV-1/EP713 cDNA clone pRFL4 (see Materials and Methods). Lane M contains DNA size markers. Lanes 2 through 7 contain nucleic acids from strain EP155 infected with CHV-1/EP713, the Δdcl2 mutant infected with CHV-1/EP713, EP155 infected with CHV-1/Euro7, the Δdcl2 mutant infected with CHV-1/Euro7, EP155 infected with CHV-1/EP721, and the Δdcl2 mutant infected with CHV-1EP721, respectively. The migration positions of genomic DNA (gDNA) and the viral double-stranded replicative-form RNA (dsRNA) and positive-sense single-stranded viral RNA (+ssRNA) are indicated on the right. The bands migrating near the 8- to 10-kb markers in lane 2 (CHV-1/EP713-infected strain EP155) are defective viral RNAs commonly observed in infected wild-type C. parasitica strains but not observed in the RNA-silencing mutant Δdcl2 strain (38). Note the significant increase in accumulation of viral positive-sense ssRNA. (B). Relative accumulation of positive-sense ssRNA hypovirus RNA in infected wild-type C. parasitica strain EP155 and Δdcl2 mutant strain determined by real-time RT-PCR. The values on the y axis were normalized to the viral RNA levels in the infected wild-type strain EP155. The relative levels of positive-sense ssRNA for CHV-1/EP713, CHV-1/Euro7, and CHV-1/EP721 increased 33-, 32-, and 16-fold in the Δdcl2 strain relative to the levels in strain EP155, respectively.