Fig 1.
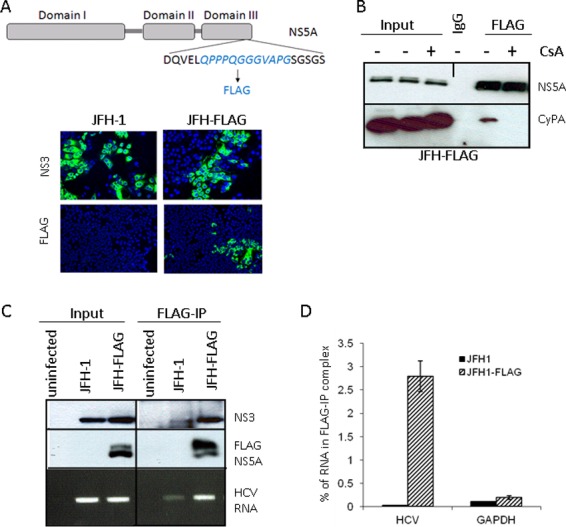
An in vivo binding assay detects NS5A-HCV RNA interaction in infected cells. (A) (Top) Schematic representation of JFH-FLAG genome. A FLAG tag (italicized) was inserted into domain III of NS5A in the JFH-1 background. (Bottom) JFH-1- and JFH-FLAG-infected cells were immunostained with an anti-NS3 (BioFront Technologies) or rabbit anti-FLAG (Sigma) antibody. (B) CyPA coprecipitates with FLAG-NS5A from infected cell lysates in a CsA-sensitive manner. CsA treatment of JFH-FLAG-infected cells was done for 18 h at a concentration of 1 μg/ml. We also included 0.1% formaldehyde in the IP mixture to stabilize the CyPA-NS5A interaction. (C) Specificity of HCV RNA pulldown from infected cell lysate by use of anti-FLAG beads. HCV RNA was detected by RT-PCR analysis of RNA immunoprecipitated from cells infected by JFH-1 or JFH-FLAG. A background band of lesser intensity that migrated slightly faster than the NS3 band was also detected in the JFH-1 sample after FLAG-IP; the identity and cause of this band are unknown. (D) Quantitative RT-PCR analysis of HCV RNA and a cellular mRNA (GAPDH) present in the FLAG-IP complex. Percentages of precipitated RNA versus input RNA were plotted.
