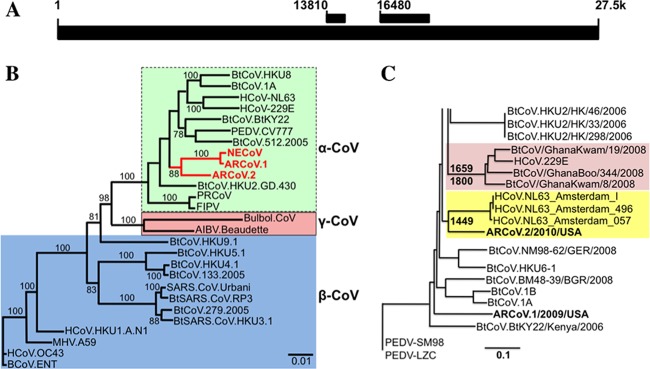
Fig 2.
A novel α-CoV in the tricolored bat is closely related to HCoV-NL63. Novel α-CoV sequences have been found in the fecal samples of several North American bat species. (A) Schematic showing where the two fragments (used for the trees in panels B and C) occur in the NL63 genome. (B) A >2.2-kb fragment of the replicase region (starting at position 16480 in the schematic) was sequenced from three samples, including two from the big brown bat (NECoV and ARCoV.1) and one from the tricolored bat (ARCoV.2). A maximum likelihood tree comparing the nucleotide sequences of these bat CoVs to other known CoVs indicated that NECoV and ARCoV.1 are very closely related while ARCoV.2 is significantly different from NECoV and ARCoV.1. The three novel α-CoV sequences form a novel cluster in the α-CoV group. (C) Molecular clock analysis using a 650- to 800-nt portion of the highly conserved replicase region (starting at position 13810 in panel A) predicted that the MRCA of bat CoVs from the Hipposideros caffer ruber bats and HCoV-229E was likely to have existed 212 to 350 years ago (in agreement with Pfefferle et al., 2009 [36]). The MRCA for HCoV-NL63 and ARCoV.2 was predicted to have existed 563 to 822 years ago. NECOV and ARCoV.1 were identical in this region, so only ARCoV.1 is shown, and it clustered with other bat α-CoVs.