Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV [human herpesvirus 8; HHV-8]) open reading frame 57 (ORF57) is a viral early protein participating in posttranscriptional regulatory events, such as splicing, RNA stabilization, and protein expression. Recent data suggest that ORF57 recruits the transcription and export (TREX) complex to viral RNA and exports these transcripts to the cytoplasm. In this study, we show that although ORF57 promotes expression of a selection of KSHV viral intronless RNAs, it is not a bona fide export factor.
TEXT
Kaposi's sarcoma-associated herpesvirus (KSHV) open reading frame 57 (ORF57) is a viral lytic protein and is essential for virus replication (3, 6, 9, 15, 20). The major roles of ORF57 occur posttranscriptionally, where it regulates splicing, stability, and expression of a subset of viral mRNAs (3, 6, 11, 12, 16, 17, 22), and whether ORF57 regulates RNA export directly remains debatable (2, 18, 21, 26). Posttranscriptional control is essential for expression of cellular and viral mRNAs, involving complex interactions of messenger RNPs (mRNPs) with transport receptors and components of the nuclear pore complex. The regulation of splicing results in the deposition of two distinct multiprotein complexes on the mRNA, namely, the exon-junction complex (EJC) and the human transcription and export (hTREX) complex (5, 23, 24, 27). Herpesviruses express several intronless RNAs requiring a regulation independent of the splicing process, and the mechanism whereby these RNAs are controlled is being elucidated. ORF57 regulates the expression of intronless viral RNAs (1–3, 16, 20); however, its exact mechanism of function remains under investigation. We recently found that ORF57 interacts with RBM15 and OTT3, two cofactors of the nuclear export receptor NXF1/TAP (10, 13, 30, 32), thereby providing a link to the export machinery (16). Importantly, this interaction is preserved among the ORF57 homologs (16), including ICP27 in herpes simplex virus 1 (HSV-1), EB2 in Epstein-Barr virus (EBV), and UL69 in human cytomegalovirus (HCMV) (1, 4, 7, 29). ORF57 was also reported to bind to intronless viral mRNAs (2, 11, 18, 22) and to recruit the hTREX complex to assemble an export-competent viral ribonucleoprotein particle via interaction of ORF57 and the Aly/REF component of the hTREX complex (2). ORF57 was implied to provide direct export function to the intronless viral mRNA (2). In the present study, we show that although KSHV ORF57 is a potent posttranscriptional regulator of selected viral RNAs, ORF57 is not a bona fide export factor.
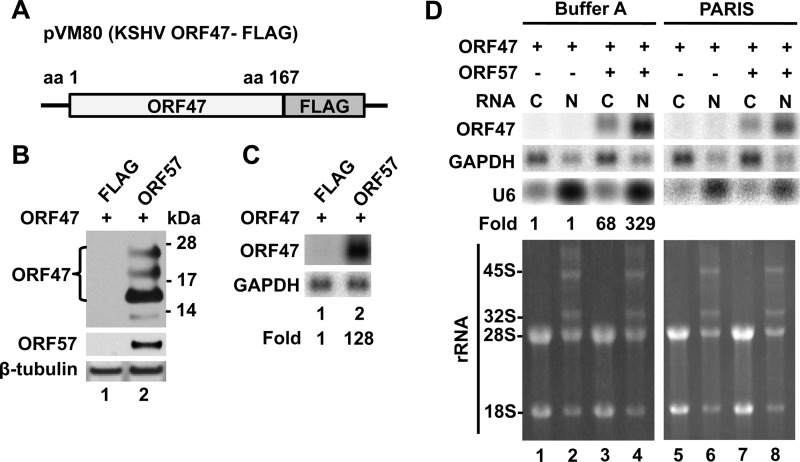
First, we examined the effect of ORF57 on the expression of KSHV ORF47 encoding viral glycoprotein L (gL). Expression of ORF47-FLAG (pVM80) (Fig. 1A) was tested in the absence and presence of the ORF57-FLAG vector (pVM7) (19) by Western blot analysis (Fig. 1B). Coexpression of ORF57 greatly increased the expression of ORF47 protein (lane 1 versus lane 2), confirming the ORF57 regulation of expression of ORF47, a glycosylated protein migrating as multiple bands.
Fig 1.
KSHV ORF57 promotes preferentially nuclear accumulation of KSHV ORF47 RNA. (A) The KSHV ORF47-FLAG fusion vector pVM80 contains the ORF47 coding region (nt 69415 to 69930; GenBank accession no. KSU75698 [28]) from Bac36-wt DNA (15) inserted into pFLAG-CMV-5.1 (Sigma, St. Louis, MO). (B and C) HEK293 cells (5 × 105 cells/well in a 6-well plate) were cotransfected with 1 μg of pVM80 and 0.5 μg of empty pFLAG vector (negative control) or ORF57-FLAG (pVM7) expression vector and harvested 24 h later in SDS-protein sample buffer with 10% 2-mercaptoethanol. The expression of ORF47-FLAG protein in the absence or presence of ORF57-FLAG was detected by Western blot analysis using anti-FLAG antibody (B). Cellular β-tubulin was used as a loading control. Northern blot analysis was used to examine ORF47 RNA expression in the absence and presence of ORF57 (C). Total RNA (5 μg/lane) isolated by TRIzol reagent (Invitrogen) was separated on an agarose gel, and ORF47 RNA was detected with the 32P-labeled oligoprobes oVM190 (nt 69770 to 69790; 5′-GCGAAGTGGATAGAGTGGACC-3′) for KSHV ORF47, oZMZ270 (5′-TGAGTCCTTCCACGATACCAAA-3′) for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and oST197 (5′-AAAATATGGAACGCTTCACGA-3′) for U6 snRNA, and fold values were normalized to GAPDH. (D) Subcellular distribution of ORF47 RNA in the presence or absence of ORF57 expression. The cytoplasmic and nuclear RNA fractions were isolated using buffer A or the Ambion PARIS kit, respectively. Five micrograms of each RNA preparation was analyzed in the Northern blot for the expression of ORF47 RNA. GAPDH served as a loading control. The fractionation efficiency was determined by the presence of U6 snRNP in the Northern blot, and 45S and 32S rRNA precursors were visualized by ethidium bromide staining.
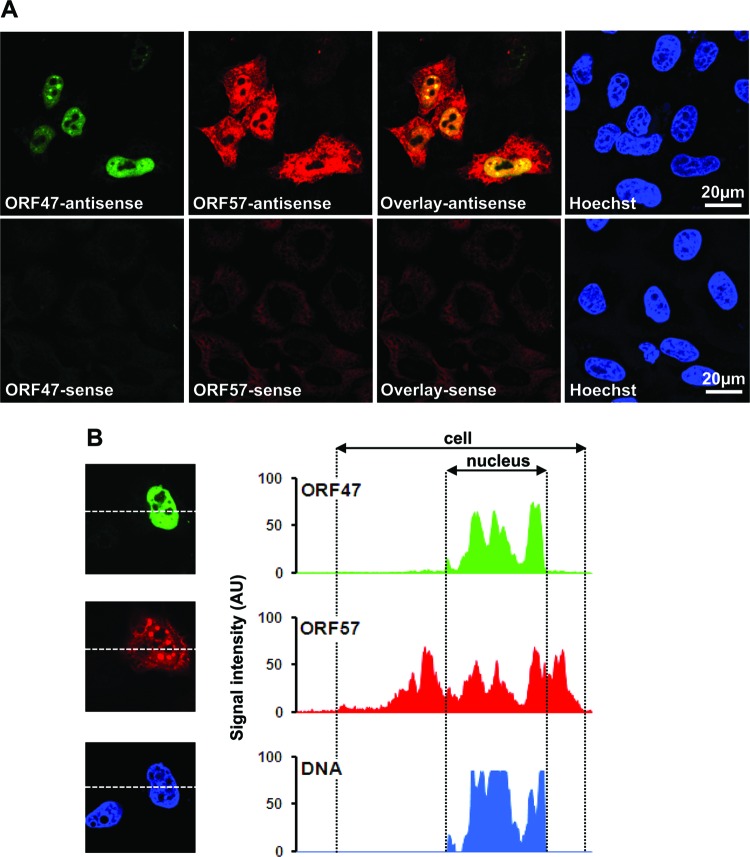
We further tested the effect of ORF57 on the overall level of the ORF47 transcript in total RNA. Northern blot analysis using an ORF47-specific probe revealed that coexpression of ORF57 resulted in ∼130-fold-increased levels of ORF47 RNA (Fig. 1C, lane 1 versus lane 2). These data are in sharp contrast with those from a previous report (2), which stated that ORF57 did not affect the level of ORF47 RNA, but only increased the export of ORF47 RNA. To test the export effect, we analyzed the subcellular levels of ORF47 RNA in the absence or presence of ORF57 by Northern blot analysis (Fig. 1D) using two different fractionation protocols: buffer A (18) in lanes 1 to 4 and the PARIS kit (2) in lanes 5 to 8. The buffer A protocol uses mild conditions (50 mM Tris [pH 80], 140 mM NaCl, 1.5 mM MgCl2, 0.2% NP-40, 200 U/ml RNasin) followed by a low-speed spin to lyse the cells and collect the cytoplasmic fraction and the nuclear pellet. Using the buffer A method, detectable levels of both cytoplasmic and nuclear ORF47 RNA were found only in the presence of ORF57 (Fig. 1D, compare lanes 1 and 3 and lanes 2 and 4), in agreement with the data shown in Fig. 1C. In the presence of ORF57, the majority of ORF47 RNA was retained in the nuclear fraction (lane 4; ∼330-fold in the nucleus). To avoid any discrepancies due to RNA fractionation with the reported data (2), we performed the same experiments using the PARIS kit as in reference 2 and obtained the same results as with buffer A (Fig. 1D, lanes 5 to 8). Under both experimental conditions in the absence of ORF57, ORF47 RNA was barely detectable. To support the subcellular fractionation data, we performed RNA fluorescent in situ hybridization (FISH) analysis on ORF47 RNA when expressed in the presence of ORF57. Similar to the Northern blot data, in the presence of ORF57 protein, ORF47 RNA was found almost exclusively in the nucleus of cells, while ORF57 RNA, which is efficiently exported as reported (16), appeared in both the nucleus and the cytoplasm (Fig. 2A). These data were confirmed by analyzing the signal intensity of a single cell, which confirmed the strong overlap in signals for ORF47 and the Hoechst nuclear stain (Fig. 2B).
Fig 2.
KSHV ORF57 does not promote RNA export of KSHV ORF47 RNA. (A) Subcellular localization of KSHV ORF47 and KSHV ORF57 transcripts determined by RNA FISH. HeLa cells (2.5 × 105 cells/well in a 6-well plate) were cotransfected by KSHV ORF47 and ORF57 expression vectors. Twenty-four hours after transfection, the cells were fixed and subcellular localizations of KSHV ORF47 (green color) and KSHV ORF57 (red color) transcripts were visualized by RNA FISH using corresponding antisense RNA probes (upper panel). The corresponding sense probes (lower panel) served as controls for probe specificity. The probes were transcribed using the FISH Tag RNA multicolor kit (Life Sciences), respectively, from a pcDNA3.0 vector-derived plasmid, pVM91 (KSHV ORF47 nt 69415 to 69930), and plasmid pVM92 (KSHV ORF57 nt 82069 to 82586). The nuclei were counterstained with Hoechst dye (blue). (B) Single-cell image of KSHV ORF47 transcripts in ORF57-coexpressing cells. A representative cell coexpressing ORF47 and ORF57 was hybridized with corresponding antisense probes as described in panel A. AU, arbitrary units. The histogram on the right shows intensity levels of each probe signal and Hoechst nucleus staining measured by Image J software along with a white dashed line crossing over the entire cell on the left.
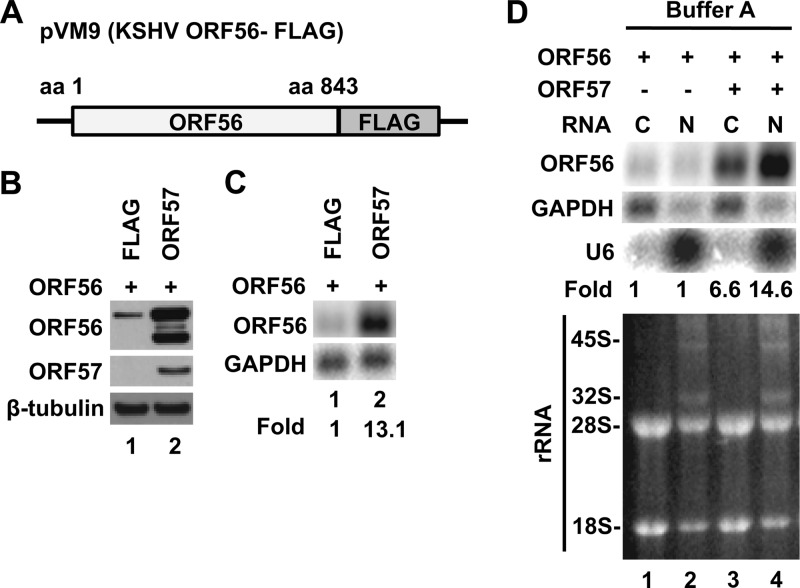
We performed similar experiments using another ORF57 target, KSHV ORF56 (Fig. 3A), encoding a viral primase. Similar to results with ORF47, ORF56 protein (Fig. 3B) and ORF56 total RNA (Fig. 3C) were only readily observed upon cotransfection with ORF57. ORF57 also promoted accumulation of ORF56 RNA in both fractions, with a greater (∼14.6-fold) increase in the nuclear fraction than in the cytoplasmic fraction (∼6.6-fold) (Fig. 3D, compare lane 3 to lane 4).
Fig 3.
KSHV ORF57 promotes preferentially nuclear accumulation of KSHV ORF56 RNA. (A) Diagram of KSHV ORF56-FLAG fusion vector pVM9. (B and C) HEK293 cells (1.5 × 106 per 60-mm dish) were cotransfected with 1,800 ng of ORF56-FLAG vector (pVM9) and 600 ng of empty vector (pFLAG, negative control) or ORF57-FLAG (pVM7) expression vector and harvested 24 h later for protein and RNA preparation. The FLAG-tagged ORF56 and ORF57 proteins were immunoblotted by anti-FLAG antibody (B). β-Tubulin served as a loading control. Faster-migrating forms of ORF56-FLAG in the presence of ORF57-FLAG most likely represent degradation products. Total RNA (5 μg/lane) was separated on an agarose gel, and ORF56 RNA was detected by Northern blotting with the 32P-labeled oligoprobe oVM24 (nt 81676 to 81654; 5′-AGTGTCGTATGCTTTATCCCAAA-3′) (C). GAPDH in the Northern blot served as a loading control for normalization of the ORF56 RNA level. (D) Effect of ORF57 on the accumulation of ORF56 RNA in cytoplasmic and nuclear fractions. The cytoplasmic and nuclear RNA fractions were isolated using buffer A. Fractionated RNAs (5 μg/lane) were analyzed by Northern blotting for the expression of ORF56 and GAPDH (loading control). U6 snRNA was detected in the Northern blot together with 45S and 32S rRNA precursors, which were visualized by ethidium bromide staining and used as an indication of fractionation efficiency.
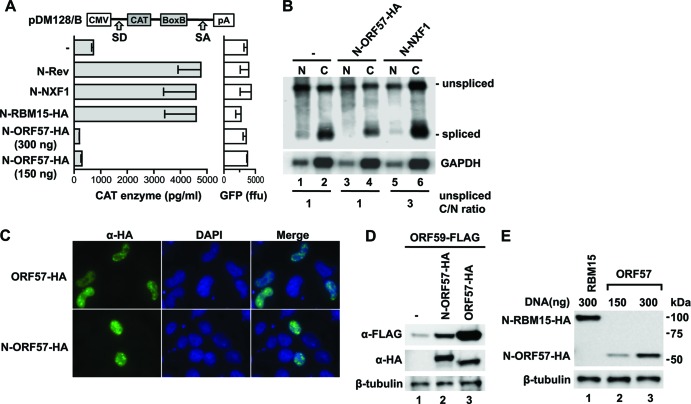
To further assess the RNA export capabilities of ORF57, we used the export reporter assay pDM128/B (31). The reporter mRNA comprises the chloramphenicol acetyltransferase (CAT) gene embedded within the HIV-1 env intronic sequence followed by six copies of the lambdoid phage p22-derived RNA-binding site (box B), serving as binding site for the phage lambda 20-amino-acid (aa) RNA-binding sequence (N-peptide) (Fig. 4A, top panel). Tethering of an export factor via the N-peptide to the reporter RNA will result in the cytoplasmic transport of the unspliced reporter RNA, resulting in CAT production. The CAT reporter plasmid was cotransfected with a plasmid expressing the fusion of the N-peptide with ORF57, and as positive controls, the previously reported plasmids expressing the fusion proteins N-Rev, N-NXF1, and N-RBM15-hemagglutinin (HA) (13, 30, 31) were included. Comparison of the levels of CAT production (Fig. 4A, bottom panel) revealed that N-Rev, N-NXF1, and N-RBM15 led to the expected export and expression of the CAT reporter RNA, while N-ORF57 (both 300 ng and 150 ng) did not promote CAT production. Northern blot analysis confirmed that N-ORF-57-HA did not affect the unspliced cat gene encoding transcript (Fig. 4B), whereas N-NXF1 increased its cytoplasmic level, as expected. This experiment also addressed ORF57's reported splicing enhancer function (14, 17, 20), which might affect the splicing of the reporter transcript and thereby remove the cat gene. The Northern blot (Fig. 4B) confirmed that the N-ORF57 fusion protein did not alter the level of the spliced CAT RNA, and hence it did not affect the splicing of the cat reporter RNA. We further confirmed that the N-peptide did not affect the localization of N-ORF57 fusion protein using fluorescence microscopy analysis and found that the N-ORF57-HA fusion protein and ORF57-HA showed similar nuclear localization (Fig. 4C). To control for a possible negative effect of the N-peptide on ORF57 function, the ability of the N-ORF57 fusion protein was examined on a known target of ORF57, KSHV ORF59 expression (18). These data showed that the N-ORF57-HA promoted expression of ORF59 and thus is functional (Fig. 4D), although we noted that the N-ORF57 fusion protein is slightly impaired (∼10-fold) in activating its natural target RNA. We further compared the expression levels of N-ORF57-HA with those of the positive control N-RBM15-HA, and Western blot analysis confirmed similar levels (Fig. 4E). Therefore, the inability of N-ORF57 to promote export and expression of the cat reporter was not due to lack of expression or function or putative splicing enhancer activity but rather was due to its lack of having a bona fide export function.
Fig 4.
KSHV ORF57 is not a bona fide export factor. (A) The diagram of pDM128/B CAT RNA shows the splice sites (SD, donor site; SA, acceptor site). HEK293 cells (1 × 106 cells per 60-mm plate) were cotransfected with 30 ng pDM128/B (31), and 300 ng of plasmids expressing the fusion proteins consisting of the phage N-peptide and the indicated export factors (N-Rev of HIV-1, N-NXF1, and N-RBM15 [13, 31]) and N-ORF57, and 48 h later, the cells were harvested and analyzed by CAT enzyme-linked immunosorbent assay (ELISA). Both N-RBM15 and N-ORF57 fusion proteins contain a C-terminal HA tag. N-ORF57-HA was expressed from an RNA-optimized N-ORF57 gene unit, and 150 and 300 ng were used. A green fluorescent protein (GFP) expression vector (100 ng pFRED25 per plate) was cotransfected to control for transfection efficiency (right panel). Raw CAT and GFP values are presented. ffu, firefly units. (B) Subcellular distribution of CAT-BoxB reporter RNA. A parallel set of plates from the HEK293 transfections (A) with 100 ng pDM128/B and 300 ng of plasmids expressing the fusion proteins was harvested for nuclear and cytoplasmic RNA isolation using the PARIS kit (Life Technologies). An equivalent amount of each RNA preparation was DNase I treated, poly(A) selected (Invitrogen), and analyzed by Northern blotting. A 32P-labeled random-primed probe was produced against the 3′ untranslated region (UTR) of the pDM128/B RNA. GAPDH served as the loading control. (C) Nuclear localization of N-peptide ORF57 fusion protein and ORF57. Plasmids (150 ng) expressing HA-tagged ORF57 and N-ORF57 were transfected into 2 × 105 HeLa cells seeded in glass-bottomed plates, fixed in 4% paraformaldehyde 1 day later, incubated with anti-HA (α-HA) primary antibody followed by Alexa Fluor 488-labeled secondary antibody, and visualized on a Zeiss Observer Z1 fluorescence microscope. DAPI, 4′,6-diamidino-2-phenylindole. (D) Function of N-ORF57 fusion protein. HEK293 cells (1 × 106 cells per 60-mm plate) were cotransfected for 24 h with ORF59-FLAG DNA (pVM18, 300 ng) with 75 ng of N-ORF57-HA and ORF57-HA DNA, respectively, and cell extracts were analyzed on Western blots. ORF59 production was detected by anti-FLAG (α-FLAG) antibody. N-ORF-57-HA and ORF57-HA expression was detected by anti-HA antibody. β-Tubulin was used as a loading control. (E) Comparison of N-ORF57-HA and N-RBM15-HA. HEK293 cells (1 × 106 cells per 60-mm plate) were transfected with the indicated plasmids, and Western blot analysis was performed by anti-HA antibody. β-Tubulin was used as a loading control.
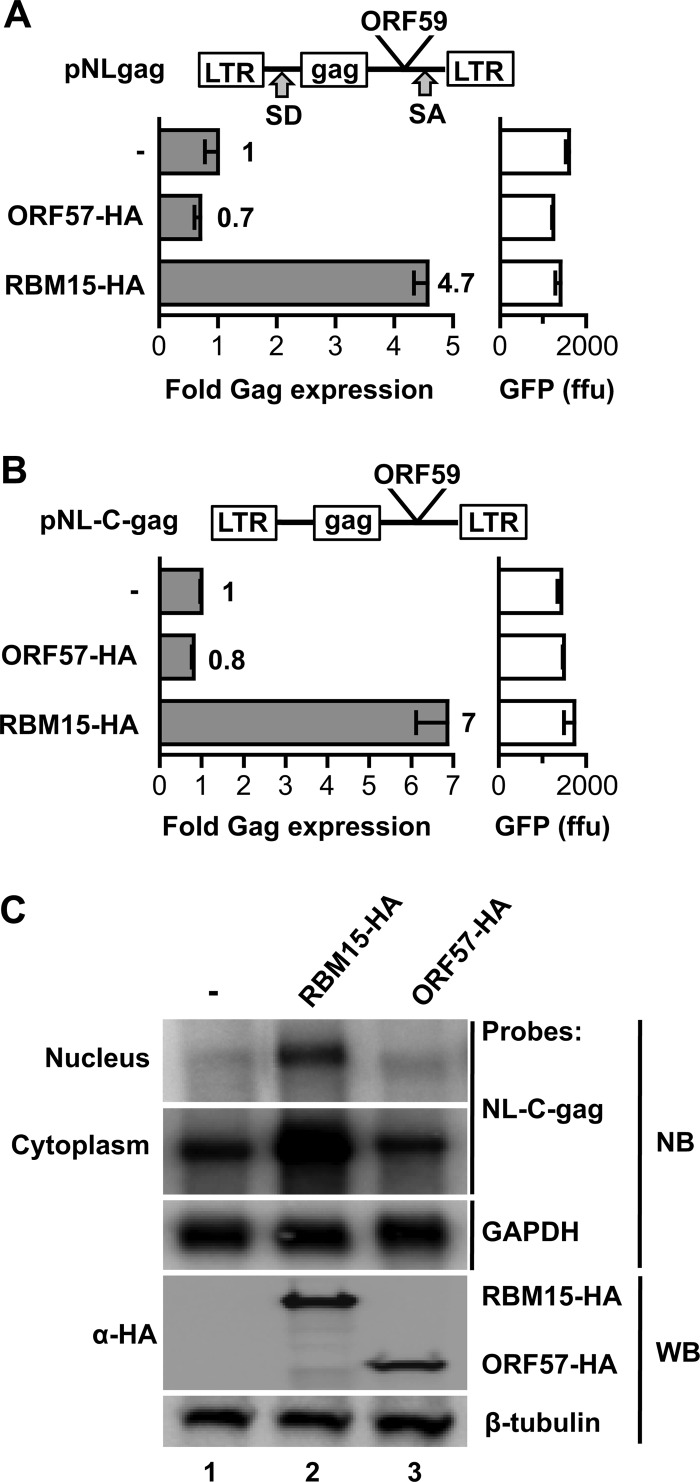
Next, we tested the export function of ORF57 using a derivative of the previously described HIV-1-based gag reporter assay (8, 25, 30) that contains the KSHV ORF59 gene (one of the reported targets of ORF57 [15, 16, 18]) inserted between the gag gene and the poly(A) signal (Fig. 5A). Gag expression was determined from the NLgag-ORF59 hybrid plasmid cotransfected in the absence or presence of plasmids expressing ORF57 or RBM15, as a positive control. Similar to the CAT reporter assay, ORF57 was unable to promote expression of the ORF59-containing Gag hybrid transcript, underscoring its lack of export function; while ectopic expression of RBM15 promoted Gag expression from the hybrid transcript, consistent with the previously reported specific activation of ORF59 expression by RBM15 (16) (Fig. 5A). Since ORF57 has been proposed to increase intronless gene expression, we also tested the expression of a nonspliceable variant produced from the NLgag-ORF59 reporter plasmid, pNL-C-gag-ORF59. Figure 5B shows that ORF57 was also unable to promote expression of the intronless gag RNA reporter, while the positive control RBM15 promoted expression of the gag reporter RNA. Subcellular fractionation of the pNL-C-gag-ORF59 RNA confirmed that ORF57 did not affect the gag RNA levels, while RBM15 increased its nuclear and cytoplasmic levels (Fig. 5C). These data show that ORF57 is unable to promote expression from either an intron-containing or non-intron-containing gag reporter transcript.
Fig 5.
KSHV ORF57 cannot export the HIV-1 gag-ORF59 chimeric RNA. (A) The KSHV ORF59 gene was cloned into the pNLgag plasmid downstream of the gag ORF, to produce pNLgagORF59. Briefly, pNLgag consists of the HIV-1 5′ long terminal repeat (LTR) and the 5′ untranslated region containing the major splice donor (SD) site of HIV-1 followed by the gag gene (nt 1 to 2621 of NL4-3 starting with the first nucleotide of U3 as +1) and the HIV-1 sequence from nt 8886 (XhoI) to the end of the 3′ LTR (nt 9709) including a cryptic splice acceptor (SA) site. (B) The KSHV ORF59 gene (without AUG and terminator of translation) was cloned into a nonspliceable pNLgag reporter plasmid (pNL-C-gag) lacking the major SD of HIV-1, downstream of gag, to produce pNL-C-gagORF59. HEK293 cells (1 × 106 cells per 60-mm plate) were cotransfected with the pNLgag-KSHV ORF59 chimeric construct (A) or pNL-C-gagORF59 (B) in the presence or absence of an ORF57-HA or RBM15-HA expression vector, and 48 h later, the cells were harvested and analyzed by HIV p24gag ELISA. Gag expression in the absence of cotransfected putative activators was normalized to 1, and the fold expression in the presence of ORF57 or RBM15 is shown. The cotransfected GFP plasmid (100 ng pFRED25 per plate) was used to measure the transfection efficiency by fluorescent measurement of the extract with a microplate reader, and the raw values are given. (C) Subcellular distribution of the NL-C-gagORF59 reporter RNA. A parallel set of plates from the HEK293 transfections (B) was harvested for nuclear and cytoplasmic RNA isolation using the PARIS kit (Life Technologies), DNase I treated, poly(A) selected (Invitrogen), and analyzed by Northern blotting. A 32P-labeled random-primed probe was produced against the NL-C-gagORF59 RNA. GAPDH RNA served as a loading control for cytoplasmic RNA. The nuclear and cytoplasmic RNA samples were run on the same gel. Western blot analysis using anti-HA antibody shows expressions of RBM15-HA and ORF57-HA, and the β-tubulin levels were visualized to confirm equal loading.
Together, several studies do not support directed RNA export function of ORF57. First, ORF57 promoted expression of KSHV nuclear noncoding PAN RNA without promoting its export (12, 15, 22). Second, despite increased cytoplasmic accumulation of the KSHV ORF59 transcript upon coexpression of ORF57, the presence or absence of ORF57 did not change the distribution of ORF59 RNA, as measured by the RNA C/N ratio (16, 18). Third, the ORF57-mediated accumulation of its target RNAs was not dependent on the hTREX core component Aly/REF (18, 26). Fourth, as shown in this report, ORF57 mediated the accumulation of intronless ORF47 and ORF56 transcripts in the nucleus as well as in the cytoplasm and promoted their expression. Fifth, ORF57 was unable to export either intron-containing or intronless reporter transcripts and thereby lacks bona fide export function. Finally, these data provide strong evidence that ORF57, although very capable of promoting expression of many KSHV genes, is not able to provide the specific RNA export function, as previously suggested (2, 21). Although not a bona fide export factor, ORF57, like its homologous proteins, plays a critical role in the posttranscriptional regulation of many viral genes and remains essential for the production of the virus.
ACKNOWLEDGMENTS
We thank T. Jones for editorial assistance.
This work was supported in part by the Intramural Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 19 September 2012
REFERENCES
- 1. Boyne JR, Colgan KJ, Whitehouse A. 2008. Herpesvirus saimiri ORF57: a post-transcriptional regulatory protein. Front. Biosci. 13:2928–2938 [DOI] [PubMed] [Google Scholar]
- 2. Boyne JR, Colgan KJ, Whitehouse A. 2008. Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog. 4:e1000194 doi:10.1371/journal.ppat.1000194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boyne JR, Jackson BR, Whitehouse A. 2010. ORF57: master regulator of KSHV mRNA biogenesis. Cell Cycle 9:2702–2703 [DOI] [PubMed] [Google Scholar]
- 4. Chen IH, Sciabica KS, Sandri-Goldin RM. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng H, et al. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127:1389–1400 [DOI] [PubMed] [Google Scholar]
- 6. Conrad NK. 2009. Posttranscriptional gene regulation in Kaposi's sarcoma-associated herpesvirus. Adv. Appl. Microbiol. 68:241–261 [DOI] [PubMed] [Google Scholar]
- 7. Farjot G, et al. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Felber BK, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis GN. 1989. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. U. S. A. 86:1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han Z, Swaminathan S. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 80:5251–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hiriart E, et al. 2005. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J. Biol. Chem. 280:36935–36945 [DOI] [PubMed] [Google Scholar]
- 11. Kang JG, et al. 2011. Kaposi's sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J. Virol. 85:2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kirshner JR, Lukac DM, Chang J, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindtner S, et al. 2006. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 281:36915–36928 [DOI] [PubMed] [Google Scholar]
- 14. Majerciak V, Kruhlak M, Dagur PK, McCoy JP, Jr, Zheng ZM. 2010. Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J. Biol. Chem. 285:11297–11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J. Virol. 81:1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Majerciak V, et al. 2011. Kaposi's sarcoma-associated herpesvirus ORF57 interacts with cellular RNA export cofactors RBM15 and OTT3 to promote expression of viral ORF59. J. Virol. 85:1528–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Majerciak V, et al. 2008. Kaposi's sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J. Virol. 82:2792–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Majerciak V, Yamanegi K, Nie SH, Zheng ZM. 2006. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 281:28365–28378 [DOI] [PubMed] [Google Scholar]
- 19. Majerciak V, Yamanegi K, Zheng ZM. 2006. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 80:11968–11981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Majerciak V, Zheng ZM. 2009. Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front. Biosci. 14:1516–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malik P, Blackbourn DJ, Clements JB. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 279:33001–33011 [DOI] [PubMed] [Google Scholar]
- 22. Massimelli MJ, et al. 2011. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5′ end to interact with viral ORF57 and cellular PABPC1. Int. J. Biol. Sci. 7:1145–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Masuda S, et al. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19:1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136:688–700 [DOI] [PubMed] [Google Scholar]
- 25. Nappi F, et al. 2001. Identification of a novel posttranscriptional regulatory element by using a rev- and RRE-mutated human immunodeficiency virus type 1 DNA proviral clone as a molecular trap. J. Virol. 75:4558–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. 2007. Kaposi's sarcoma-associated herpesvirus ORF57 protein enhances mRNA accumulation independently of effects on nuclear RNA export. J. Virol. 81:9990–9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reed R, Cheng H. 2005. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 17:269–273 [DOI] [PubMed] [Google Scholar]
- 28. Russo JJ, et al. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Toth Z, Stamminger T. 2008. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front. Biosci. 13:2939–2949 [DOI] [PubMed] [Google Scholar]
- 30. Uranishi H, et al. 2009. The RNA-binding motif protein 15B (RBM15B/OTT3) acts as cofactor of the nuclear export receptor NXF1. J. Biol. Chem. 284:26106–26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wiegand HL, Lu S, Cullen BR. 2003. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. U. S. A. 100:11327–11332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zolotukhin AS, et al. 2009. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. 37:7151–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]