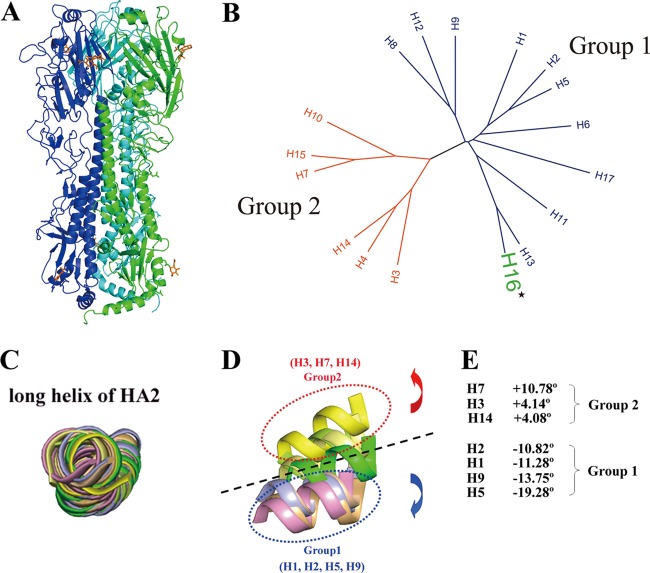
Fig 1.
Overall structure of H16H0 and HA grouping based on 3D structures. (A) Overview of the H16HA0 trimer, represented as a ribbon diagram. For clarity, each monomer has been colored differently (A, green; B, blue; C, cyan). Carbohydrates observed in the electron density maps are colored orange. (B) Phylogenetic tree of the 17 HA subtypes. The 17 HA subtypes can be divided phylogenetically into two groups on the basis of their full-length sequences: group 1 (blue) and group 2 (red). H16, highlighted in green, belongs to group 1. (C to E) Superimpositions of the long α helix (residues 76 to 126) of HA2 reveal the displacements of the HA1 globular subdomain in seven HA subtype structures. The 190 helix (residues 188 to 195) in the receptor binding domain is used as a representative of the globular subdomain of each HA subtype. H1 is colored light blue, H2 is wheat, H3 is lemon, H5 is light orange, H7 is yellow, H9 is pink, H14 is pale yellow, and H16 is green. When we use H16 as a reference, the group 1 members rotate downward with negative-angle degrees (from −10.82° to −19.28°), while the group 2 members rotate upward with positive-angle degrees (from 4.08° to 10.78°).