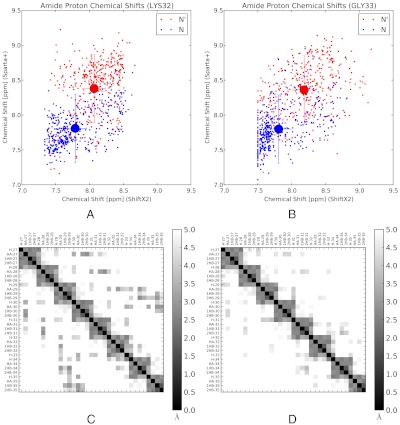
Fig. 5.
(A, B). Amide proton chemical shifts were calculated for 400 randomly selected conformations from the native and near-native states of HP35. Calculations were performed with both ShiftX2 and Sparta+. The large symbol represents the in-state means of the native and near-native chemical shifts. Error bars correspond to the RMS uncertainty estimated in the parameterization of the ShiftX2 and Sparta+ algorithms; compared to this systematic uncertainty, the statistical uncertainty (SEM) is insignificant. Finally, note that the predictions of ShiftX2 and Sparta+ are not strongly correlated (r2 ≈ 0.3), so repeating the calculations with both models gives greater confidence in the results. (C, D). NOE-weighted average distance (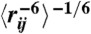 ) is shown for the N (C) and N’ (D) states. Distances are shown for residues 27–35; residues 1–26 show insignificant differences between states N and N’.
) is shown for the N (C) and N’ (D) states. Distances are shown for residues 27–35; residues 1–26 show insignificant differences between states N and N’.