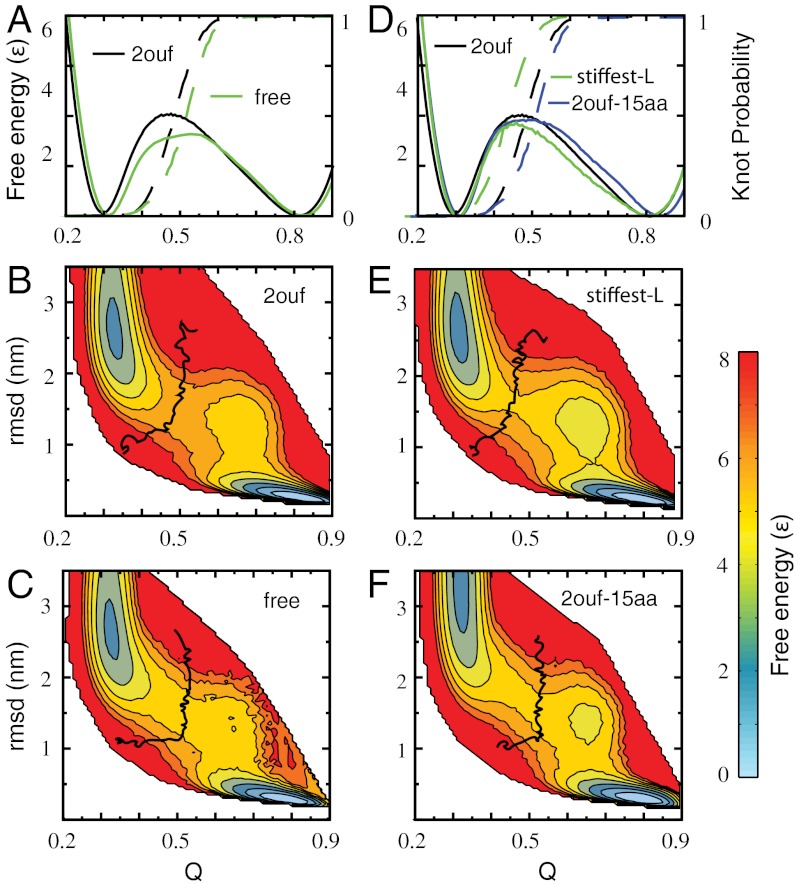
Fig. 2.
Free energy landscape of knotted proteins measured by the fraction of native contacts formed Q and rmsd for 2ouf and three mutants. (A) One-dimensional free energy F(Q) for 2ouf with 2ouf-free, and corresponding knot probability K(Q). (B and C) Two-dimensional free energy landscape of 2ouf and 2ouf-free. (D) One-dimensional free energy F(Q) for 2ouf-stiffer-L and 2ouf-15aa (longest linker). The 2ouf-stiffer-L shows the slowest knotting kinetics (Fig. 4). (E and F) Two-dimensional free energy landscape of 2ouf-stiffer-L and 2ouf-15aa. All F(Q, rmsd) show that the folding mechanism of knotted protein is composed of complex events, as seen by the chair-like shapes of the transition states and the metastable states around Q = 0.75 and rmsd = 1.2. Black curve on (B, C, E, F) shows the contour of knot probability K(Q,rmsd) = 0.5. This contour lies across the transitions state, thus coinciding with the rate-limiting step to folding. (B and E) When the contacts between the linker and the rest of the protein are included K(Q,rmsd) has the gradually curved “)” shape. (C and F) Proteins with free linkers are characterized by “⌋” shape contours. F(Q) and F(Q,RMSD) for other constructs is shown in SI Appendix, Fig. S2.