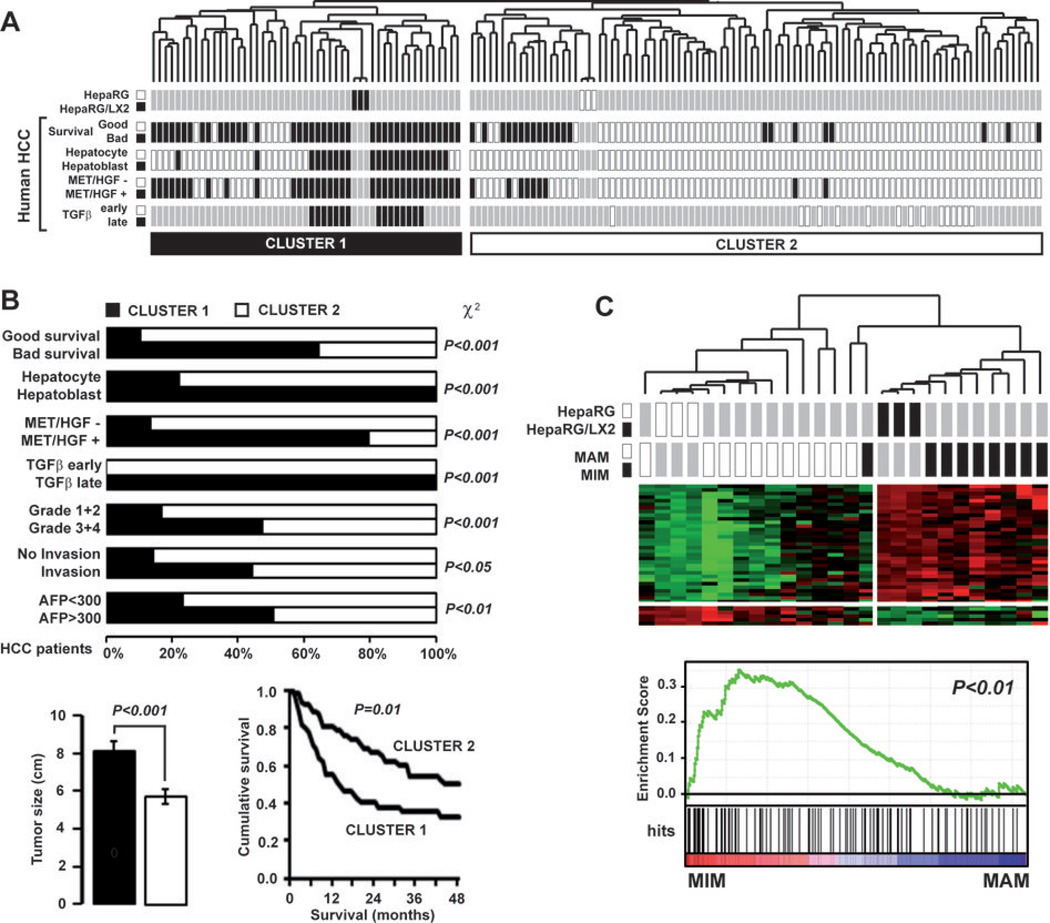
Figure 5.
Clinical relevance of HepaRG/LX2 signature in human HCC. (A) Dendrogram overview of HepaRG and HepaRG/LX2 experiments integrated with 139 cases of human HCC. Clustering analysis was based on the expression of 212 genes differentially expressed in HepaRG cocultured with LX2. Two major clusters (1 and 2) were identified. Distribution of human HCC samples between previously described subgroups with respect to survival (27) (good vs bad prognosis), cell origin (28) (hepatoblast vs hepatocytes), activation of MET/HGF (26) (− vs +) and TGFβ signaling pathway (20) (early vs late) is indicated on the left. (B) Statistical analysis of HCC distribution between clusters 1 and 2 based on previous gene signatures and clinical parameters. Cluster 1, which is defined by the HepaRG/LX2 coculture signature, shows a significant enrichment in HCC with the following features: bad survival, hepatoblast traits, activation of MET/HGF and late TGFβ pathways, higher differentiation grade and serum AFP level. Tumors size was significantly higher for HCC included in cluster 1. Kaplan-Meier plots and log-rank statistics analysis revealed a significant decreased in overall survival for patients included in cluster 1. (C) Integrative genomics using HepaRG/LX2 signature and gene expression profiles of peri-tumoral cirrhotic tissues from patient with (MIM) or without (MAM) metastasis (31). Clustering analysis (upper panel) and GSEA (lower panel) shows that the HepaRG/LX2 signature was significantly enriched in the gene expression profiles of cirrhotic tissues from patients with metastasis (MIM).