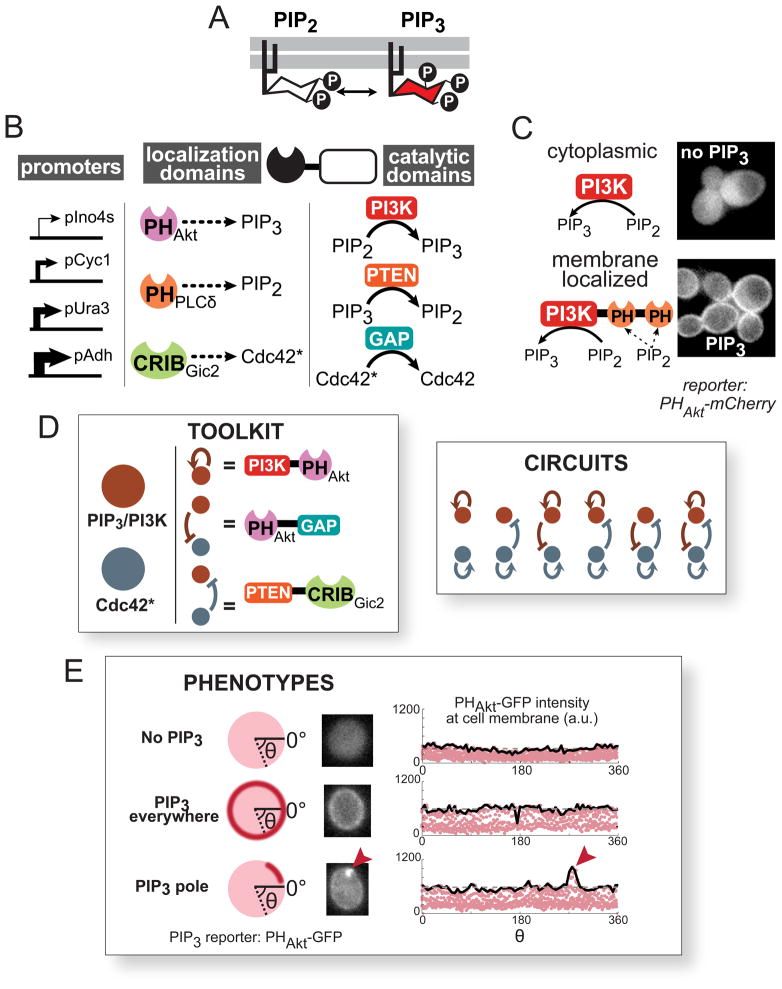
Figure 4. Construction of a synthetic polarization system using PIP3, a phospholipid not normally found in S. cerevisiae.
(A) Phosphatidylinositol (4,5)-bisphosphate (PIP2) can be reversibly phosphorylated to become PIP3. (B) Modular localization domains can be fused to catalytic domains to direct enzyme activity to specific subcellular locations. Pleckstrin homology (PH) domains bind specific phospholipid headgroups, and the CRIB (Cdc42/Rac interactive binding) domain of Gic2 localizes to activated Cdc42 (herein denoted as Cdc42*). Catalytic domains were truncated or mutated to achieve localization-dependent activity. Expression was tuned using a set of different strength constitutive yeast promoters. (C) PI3K does not produce PIP3 unless localized by the PH domain of PLCδ (PHPLCδ) to its substrate, PIP2, at the plasma membrane. The PH domain of Akt (PHAkt) fused to mCherry is used as a reporter for PIP3. See also Figure S4. (D) Using the localization and catalytic domains described above, a toolkit of enzymatic fusions is created analogous to the regulatory links in the computational model. Using this toolkit, we experimentally tested a subset of the predicted core network topologies and determined the resulting phenotypes. (E) Using 2xPHAkt-2xGFP as a reporter, we classify cells into three phenotypes: no visible PIP3, PIP3 everywhere on the plasma membrane, and a localized pole of PIP3 (see Extended Experimental Procedures). Fluorescence images of individual cells are cropped to contain only one cell, and pixel intensities at the cell membrane are plotted (red dots, right). The maximum intensity along the cell membrane (solid black line) is used to calculate the polarity score (See Figure S4). A cell without PIP3 (top) displays uniform, low intensity background fluorescence along its periphery. A cell with PIP3 everywhere (middle) displays uniform high intensity fluorescence along its edge. A cell with a PIP3 pole (bottom) has a fluorescence intensity peak indicating a local concentration of PIP3 (red arrow).