Table 1.
Strategy for identifying amino acid residues modified by ubiquitin and Ubls. The conjugated proteins are usually digested by trypsin to generate a specific tag (e.g., GG for ubiquitin) on the modified residues, resulting in a mass shift that is detectable during MS and MS/MS scans. The mass of human Fat10 tag may change because of natural variations in sequence (UniProtKB/Swiss-Prot O15205).
| Human Ub or UBL | Remnant on modified peptides after trypsin digestion | Mass change in MS and MS/MS | Problems | Solutions |
|---|---|---|---|---|
| Ub |
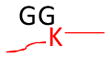
|
Monoisotopic mass shift of 114.0429 Da in MS and MS/MS | Ub, Nedd8, and ISG15 modifications lead to the same mass change after trypsin digestion | Pre-enrichment of modified proteins prior totrypsin digestion |
| Nedd8 |
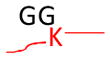
|
|||
| ISG15 |
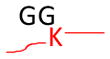
|
|||
| SUMO-1 |
|
Large mass shift in MS and complex production patterns in MS/MS | Weak signal in MS and complex MS/MS spectra that are difficult to interpret |
|
| SUMO-2/3 |
|
|||
| Fat10 |
|
